Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III - PubMed (original) (raw)
Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III
M Ahmed et al. J Virol. 1998 Oct.
Abstract
The matrix (M) protein of vesicular stomatitis virus (VSV) functions in virus assembly and inhibits host-directed gene expression independently of other viral components. Experiments in this study were carried out to determine the ability of M protein to inhibit transcription directed by each of the three host RNA polymerases (RNA polymerase I [RNAPI], RNAPII, and RNAPIII). The effects of wild-type (wt) VSV, v6 (a VSV mutant isolated from persistently infected cells), and tsO82 viruses on poly(A)+ and poly(A)- RNA synthesis were measured by incorporation of [3H]uridine. v6 and tsO82 viruses, which contain M-gene mutations, had a decreased ability to inhibit synthesis of both poly(A)+ and poly(A)- RNA. Nuclear runoff analysis showed that VSV inhibited transcription of 18S rRNA and alpha-tubulin genes, which was dependent on RNAPI and RNAPII, respectively, but infection with wt virus enhanced transcription of 5S rRNA by RNAPIII. The effect of M protein alone on transcription by RNAPI-, RNAPII-, and RNAPIII-dependent promoters was measured by cotransfection assays. M protein inhibited transcription from RNAPI- and RNAPII-dependent promoters in the absence of other viral gene products. RNAPIII-dependent transcription of the adenovirus VA promoters was also inhibited by M protein. However, as observed during wt VSV infection, M protein enhanced endogenous 5S rRNA transcription, indicating that the inhibition of transcription by RNAPIII was dependent on the nature of the promoter.
Figures
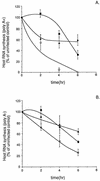
FIG. 1
Poly(A)+ (A) and poly(A)− (B) RNA synthesis in BHK cells infected with wt, _ts_O82, and v6 viruses. BHK cells were infected with wt (open circles), _ts_O82 (closed squares), and v6 (closed triangles) viruses at a multiplicity of infection of 20 PFU/cell. Parallel samples were infected in the presence of actinomycin D. At 2, 4, and 6 h postinfection, cells were labeled with [3H]uridine (20 μCi/ml) for 30 min. Cells were harvested and lysed in SDS lysis buffer. To separate poly(A)+ and poly(A)− RNAs, lysates were incubated in the presence of oligo(dT) cellulose (Invitrogen), washed in high-salt buffer, and eluted in low-salt buffer. Samples were then precipitated with 7% trichloroacetic acid on ice and washed twice with 7% trichloroacetic acid. Acid-precipitable radioactivity was measured by scintillation counting. Values of samples incubated in the presence of actinomycin D were subtracted from the total counts to determine the rate of host RNA synthesis. Data shown are means ± standard deviations for four experiments.
FIG. 2
Effect of M protein on transcriptional activity of genes dependent on RNAPI. (A) BHK cells were cotransfected with pHrMr plasmid DNA and 0, 36, or 360 ng of in vitro-transcribed wt M mRNA. Cells that received no M mRNA were cotransfected with 360 ng of yeast RNA as a negative control. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized pHrMr plasmid DNA fixed on nitrocellulose membrane filters. (B) BHK cells were cotransfected with pHrMr plasmid DNA and 0 or 360 ng of _ts_O82 M mRNA. Transcription of pHrMr DNA was assayed by nuclear runoff analysis as described above for panel A. (C) BHK cells were transfected with pHrMr DNA or no plasmid DNA as a control for the specificity of hybridization. Transcription of pHrMr DNA was assayed by nuclear runoff analysis as described above for panel A. (D) BHK cells were infected at a multiplicity of infection of 20 PFU/cell with wt or _ts_O82 virus. Mock-infected cells were used as a control. Nuclei were isolated 6 h postinfection, and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to a cDNA fragment of 18S rRNA immobilized on nitrocellulose membranes. (E) The data from four (A and D) or three (B) separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA for the transfection experiments and as a percentage of the uninfected control for the virus-infected cells. The data are means ± standard deviations.
FIG. 3
Effect of M protein on transcriptional activity of genes dependent on RNAPII. (A) BHK cells were cotransfected with pSV2.CAT plasmid DNA and 0, 36, or 360 ng of in vitro-transcribed wt M mRNA. Cells that received no M mRNA were cotransfected with 360 ng of yeast RNA as a negative control. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized pSV2.CAT plasmid DNA fixed on nitrocellulose membrane filters. (B) BHK cells were cotransfected with pSV2.CAT plasmid DNA and 0 or 360 ng of _ts_O82 M mRNA. Transcription of pSV2.CAT DNA was assayed by nuclear runoff analysis as described above for panel A. (C) BHK cells were transfected with pSV2.CAT DNA or no plasmid DNA as a control for the specificity of hybridization. Transcription of pSV2.CAT DNA was assayed by nuclear runoff analysis as described above for panel A. (D) BHK cells were infected at a multiplicity of infection of 20 PFU/cell with wt or _ts_O82 virus. Mock-infected cells were used as a control. Nuclei were isolated 6 h postinfection, and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to a cDNA fragment of α-tubulin mRNA immobilized on nitrocellulose membranes. (E) The data from four (A and B) or two (D) separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA for the transfection experiments and as a percentage of the uninfected control in the case of the virus-infected cells. The data are means ± standard deviations.
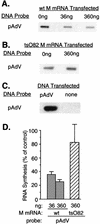
FIG. 4
Effect of M protein on transcriptional activity of adenovirus VA genes dependent on RNAPIII. (A) BHK cells were cotransfected with pAdVantage (pAdV) plasmid DNA and 0, 36 or 360 ng of in vitro-transcribed wt M mRNA. Cells that received no M mRNA were cotransfected with 360 ng of yeast RNA as a negative control. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized pAdV plasmid DNA fixed on nitrocellulose membrane filters. (B) BHK cells were cotransfected with pAdV plasmid DNA and 0 or 360 ng of _ts_O82 M mRNA. Transcription of pAdV DNA was assayed by nuclear runoff analysis as described above for panel A. (C) BHK cells were transfected with pAdV DNA or no plasmid DNA as a control for the specificity of hybridization. Transcription of pAdVantage DNA was assayed by nuclear runoff analysis as described above for panel A. (D) The data from four (A) or three (B) separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA. The data are means ± standard deviations.
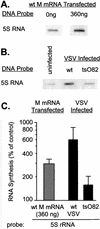
FIG. 5
Effect of M protein on transcriptional activity of 5S rRNA genes dependent on RNAPIII. (A) BHK cells were transfected with 0 or 360 ng of in vitro-transcribed wt M mRNA. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized cDNA of 5S rRNA fixed on nitrocellulose membrane filters. (B) BHK cells were infected at a multiplicity of infection of 20 PFU/cell with wt or _ts_O82 virus. Mock-infected cells were used as a control. Nuclei were isolated 6 h postinfection, and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to a cDNA of 5S rRNA immobilized on nitrocellulose membranes. (C) The data from four separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA for the transfection experiments and as a percentage of the uninfected control in the case of the virus-infected cells. The data are means ± standard deviations and are plotted on a logarithmic scale to accommodate all of the values.
Similar articles
- Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis.
Ahmed M, McKenzie MO, Puckett S, Hojnacki M, Poliquin L, Lyles DS. Ahmed M, et al. J Virol. 2003 Apr;77(8):4646-57. doi: 10.1128/jvi.77.8.4646-4657.2003. J Virol. 2003. PMID: 12663771 Free PMC article. - The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter.
Ferran MC, Lucas-Lenard JM. Ferran MC, et al. J Virol. 1997 Jan;71(1):371-7. doi: 10.1128/JVI.71.1.371-377.1997. J Virol. 1997. PMID: 8985359 Free PMC article. - VSV RNA synthesis: how can you be positive?
Ball LA, Wertz GW. Ball LA, et al. Cell. 1981 Oct;26(2 Pt 2):143-4. doi: 10.1016/0092-8674(81)90297-x. Cell. 1981. PMID: 6277494 Review. No abstract available. - Evolution of viral DNA-dependent RNA polymerases.
Sonntag KC, Darai G. Sonntag KC, et al. Virus Genes. 1995;11(2-3):271-84. doi: 10.1007/BF01728665. Virus Genes. 1995. PMID: 8828152 Review.
Cited by
- High-throughput virtual screening and docking studies of matrix protein vp40 of ebola virus.
Tamilvanan T, Hopper W. Tamilvanan T, et al. Bioinformation. 2013;9(6):286-92. doi: 10.6026/97320630009286. Epub 2013 Mar 19. Bioinformation. 2013. PMID: 23559747 Free PMC article. - RNA interference with measles virus N, P, and L mRNAs efficiently prevents and with matrix protein mRNA enhances viral transcription.
Reuter T, Weissbrich B, Schneider-Schaulies S, Schneider-Schaulies J. Reuter T, et al. J Virol. 2006 Jun;80(12):5951-7. doi: 10.1128/JVI.02453-05. J Virol. 2006. PMID: 16731933 Free PMC article. - Vesicular stomatitis virus matrix protein impairs CD1d-mediated antigen presentation through activation of the p38 MAPK pathway.
Renukaradhya GJ, Khan MA, Shaji D, Brutkiewicz RR. Renukaradhya GJ, et al. J Virol. 2008 Dec;82(24):12535-42. doi: 10.1128/JVI.00881-08. Epub 2008 Sep 24. J Virol. 2008. PMID: 18815300 Free PMC article. - Early steps of the virus replication cycle are inhibited in prostate cancer cells resistant to oncolytic vesicular stomatitis virus.
Carey BL, Ahmed M, Puckett S, Lyles DS. Carey BL, et al. J Virol. 2008 Dec;82(24):12104-15. doi: 10.1128/JVI.01508-08. Epub 2008 Oct 1. J Virol. 2008. PMID: 18829743 Free PMC article. - Crosstalk between nucleocytoplasmic trafficking and the innate immune response to viral infection.
Shen Q, Wang YE, Palazzo AF. Shen Q, et al. J Biol Chem. 2021 Jul;297(1):100856. doi: 10.1016/j.jbc.2021.100856. Epub 2021 Jun 29. J Biol Chem. 2021. PMID: 34097873 Free PMC article. Review.
References
- Ahmed M, Lyles D S. Identification of a consensus mutation in M protein of vesicular stomatitis virus from persistently infected cells that affects inhibition of host-directed gene expression. Virology. 1997;237:378–388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources