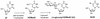Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei - PubMed (original) (raw)
Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei
F van Leeuwen et al. Mol Cell Biol. 1998 Oct.
Abstract
beta-D-Glucosyl-hydroxymethyluracil, also called J, is a modified DNA base conserved among kinetoplastid flagellates. In Trypanosoma brucei, the majority of J is present in repetitive DNA but the partial replacement of thymine by J also correlates with transcriptional repression of the variant surface glycoprotein (VSG) genes in the telomeric VSG gene expression sites. To gain a better understanding of the function of J, we studied its biosynthesis in T. brucei and found that it is made in two steps. In the first step, thymine in DNA is converted into hydroxymethyluracil by an enzyme that recognizes specific DNA sequences and/or structures. In the second step, hydroxymethyluracil is glucosylated by an enzyme that shows no obvious sequence specificity. We identified analogs of thymidine that affect the J content of the T. brucei genome upon incorporation into DNA. These analogs were used to study the function of J in the control of VSG gene expression sites. We found that incorporation of bromodeoxyuridine resulted in a 12-fold decrease in J content and caused a partial derepression of silent VSG gene expression site promoters, suggesting that J might strengthen transcriptional repression. Incorporation of hydroxymethyldeoxyuridine, resulting in a 15-fold increase in the J content, caused a reduction in the occurrence of chromosome breakage events sometimes associated with transcriptional switching between VSG gene expression sites in vitro. We speculate that these effects are mediated by the packaging of J-containing DNA into a condensed chromatin structure.
Figures
FIG. 1
Putative biosynthetic pathway for J. First, a thymidine (dT) residue in a certain context in DNA is converted into HOMedU by a DNA thymidine-7-hydroxylase. Second, HOMedU in DNA is converted into β-
d
-glucosyl-HOMedU (dJ) by a β-glucosyl transferase. BrdU is a thymidine analog that cannot be converted into dJ.
FIG. 2
Analysis of thymidine analogs incorporated into trypanosome DNA by 32P-nucleotide postlabeling combined with 2D-TLC. (A) Schematic representation of the positions of the deoxynucleotides (solid circles), including dJ (large arrow), HOMedU (small arrow), and BrdU (arrowhead). The ribonucleotides (open circles) and unknown products (dashed open circles) that contaminate the DNA, enzyme, and label preparations varied per experiment or batch; ribonucleotides were more abundant in small-scale DNA preparations (E and F). Trypanosomes were grown in the presence or absence of thymidine analogs. (B to F) Chromatograms representing WT PF trypanosomes (B), TKN PF trypanosomes plus 1 mM HOMedU (C), WT BF trypanosomes (D), HTK BF trypanosomes plus 1 mM HOMedU (E), and HTK BF trypanosomes plus 100 μM BrdU (F). In the absence of nucleoside analogs, WT trypanosomes and TK transformants have the same nucleotide composition (data not shown).
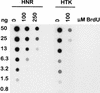
FIG. 3
Anti-J DNA dot blot analysis. DNA of HNR and HTK BF trypanosomes (see Fig. 4 and 5) grown in the absence or presence of BrdU was loaded as a twofold dilution series onto a dot blot and incubated with rabbit anti-J antiserum. Bound antibody was detected with a sheep anti-rabbit secondary antibody conjugated to horseradish peroxidase and visualized by enhanced chemiluminescence. We have previously shown that the detection of J on immunoblots is not affected by the presence of nonmodified DNA (56). A twofold decrease in the enhanced chemiluminescence signal therefore corresponds to a twofold increase in the J content if equal amounts of DNA are loaded. After the antibodies were stripped off, DNA loading was checked by hybridization with a RIME probe (results not shown; see Materials and Methods).
FIG. 4
Northern blot analysis of RNA from BF trypanosomes cultivated in the absence or presence of thymidine analogs. (A) BF clones HNR and HN1, which are genotypically identical but have a different active expression site (15), and clone RP2XR (47) are described in Materials and Methods. A solid flag indicates an endogenous expression site promoter, and an open flag indicates a ribosomal promoter. Transcription is indicated by a dashed line with arrowhead, and the vertical line downstream of the VSG genes indicates the chromosome end. (B) Northern blots of HN1 control cells and of HNR and BF trypanosomes grown in the absence or presence of BrdU were hybridized with the probes indicated on the left. TUB indicates β-tubulin genes. (C) Northern blots of HN1 control cells and of HNR and RP2XR trypanosomes grown in the absence or presence of HOMedU.
FIG. 5
Effect of thymidine analogs on VSG gene expression site switching in vitro. (A) HTK cells contain HYG and TK genes downstream of the active 221 expression site promoter (flag) and are resistant to hygromycin and sensitive to BVDU (HygR, BVDUS). The dashed line with the arrowhead indicates transcription. Following negative selection, the switched trypanosome clones, which are hygromycin sensitive and BVDU resistant (HygS, BVDUR), were analyzed by DNA dot blot hybridization for the absence (−) or presence (+) of the HYG and TK (HT) genes and the VSG 221 gene (221). On the basis of the dot blot hybridization, two genotypes of expression site switch variants could be distinguished (see the text): variants that had retained the old expression site (HT/221+) and variants that had deleted completely the old expression site and thereby lost the marker genes and the VSG gene (HT/221−). (B and C) HTK cells were grown in the absence (−) or presence (+) of 100 μM BrdU (B) or 1 mM HOMedU (C). The relative number of switchers of each genotype is indicated on the y axis as a percentage of the total number of switched clones in the untreated control population of each panel (HT/221− plus HT/221+ = 100%). For each growth condition, two to five independent HTK cultures were put through a switch experiment, and the data shown in panels B and C represent the means and standard deviations of the switch patterns found. In total, we analyzed 44 clones (n = 3) and 54 clones (n = 5) for growth in the absence and presence of BrdU, respectively, and 14 clones (n = 2) and 68 clones (n = 5) for growth in the absence and presence of HOMedU, respectively. (n indicates the number of cultures used for each condition.) One clone with an H+T−221− genotype was found in the −BrdU control cells (not included in the diagram).
Similar articles
- Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei.
van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, Borst P. van Leeuwen F, et al. Genes Dev. 1997 Dec 1;11(23):3232-41. doi: 10.1101/gad.11.23.3232. Genes Dev. 1997. PMID: 9389654 Free PMC article. - beta-D-glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida.
Borst P, van Leeuwen F. Borst P, et al. Mol Biochem Parasitol. 1997 Dec 1;90(1):1-8. doi: 10.1016/s0166-6851(97)00170-9. Mol Biochem Parasitol. 1997. PMID: 9497027 Review. - Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei.
van Leeuwen F, Kieft R, Cross M, Borst P. van Leeuwen F, et al. Mol Biochem Parasitol. 2000 Jul;109(2):133-45. doi: 10.1016/s0166-6851(00)00247-4. Mol Biochem Parasitol. 2000. PMID: 10960172 - Identification of the glucosyltransferase that converts hydroxymethyluracil to base J in the trypanosomatid genome.
Bullard W, Lopes da Rosa-Spiegler J, Liu S, Wang Y, Sabatini R. Bullard W, et al. J Biol Chem. 2014 Jul 18;289(29):20273-82. doi: 10.1074/jbc.M114.579821. Epub 2014 Jun 2. J Biol Chem. 2014. PMID: 24891501 Free PMC article. - Base J: discovery, biosynthesis, and possible functions.
Borst P, Sabatini R. Borst P, et al. Annu Rev Microbiol. 2008;62:235-51. doi: 10.1146/annurev.micro.62.081307.162750. Annu Rev Microbiol. 2008. PMID: 18729733 Review.
Cited by
- G-Quadruplexes as Key Transcriptional Regulators in Neglected Trypanosomatid Parasites.
Monti L, Di Antonio M. Monti L, et al. Chembiochem. 2023 Jun 15;24(12):e202300265. doi: 10.1002/cbic.202300265. Epub 2023 May 23. Chembiochem. 2023. PMID: 37146230 Free PMC article. Review. - Advances in Protozoan Epigenetic Targets and Their Inhibitors for the Development of New Potential Drugs.
Gaona-López C, Vazquez-Jimenez LK, Gonzalez-Gonzalez A, Delgado-Maldonado T, Ortiz-Pérez E, Nogueda-Torres B, Moreno-Rodríguez A, Vázquez K, Saavedra E, Rivera G. Gaona-López C, et al. Pharmaceuticals (Basel). 2023 Apr 4;16(4):543. doi: 10.3390/ph16040543. Pharmaceuticals (Basel). 2023. PMID: 37111300 Free PMC article. Review. - Behind Base J: The Roles of JBP1 and JBP2 on Trypanosomatids.
Assis LHC, de Paiva SC, Cano MIN. Assis LHC, et al. Pathogens. 2023 Mar 16;12(3):467. doi: 10.3390/pathogens12030467. Pathogens. 2023. PMID: 36986389 Free PMC article. Review. - Chromatin-Associated Protein Complexes Link DNA Base J and Transcription Termination in Leishmania.
Jensen BC, Phan IQ, McDonald JR, Sur A, Gillespie MA, Ranish JA, Parsons M, Myler PJ. Jensen BC, et al. mSphere. 2021 Feb 24;6(1):e01204-20. doi: 10.1128/mSphere.01204-20. mSphere. 2021. PMID: 33627513 Free PMC article. - Emergence of a novel immune-evasion strategy from an ancestral protein fold in bacteriophage Mu.
Karambelkar S, Udupa S, Gowthami VN, Ramachandra SG, Swapna G, Nagaraja V. Karambelkar S, et al. Nucleic Acids Res. 2020 Jun 4;48(10):5294-5305. doi: 10.1093/nar/gkaa319. Nucleic Acids Res. 2020. PMID: 32369169 Free PMC article.
References
- Barry J D. The relative significance of mechanisms of antigenic variation in African trypanosomes. Parasitol Today. 1997;13:212–218. - PubMed
- Bernards A, de Lange T, Michels P A, Liu A Y, Huisman M J, Borst P. Two modes of activation of a single surface antigen gene of Trypanosoma brucei. Cell. 1984;36:163–170. - PubMed
- Bernards A, Van der Ploeg L H, Frasch A C, Borst P, Boothroyd J C, Coleman S, Cross G A. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. - PubMed
- Blundell P A, Rudenko G, Borst P. Targeting of exogenous DNA into Trypanosoma brucei requires a high degree of homology between donor and target DNA. Mol Biochem Parasitol. 1996;76:215–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources