The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene - PubMed (original) (raw)
The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene
P Honkakoski et al. Mol Cell Biol. 1998 Oct.
Abstract
PBREM, the phenobarbital-responsive enhancer module of the cytochrome P-450 Cyp2b10 gene, contains two potential nuclear receptor binding sites, NR1 and NR2. Consistent with the finding that anti-retinoid X receptor (RXR) could supershift the NR1-nuclear protein complex, DNA affinity chromatography with NR1 oligonucleotides enriched the nuclear orphan receptor RXR from the hepatic nuclear extracts of phenobarbital-treated mice. In addition to RXR, the nuclear orphan receptor CAR was present in the same enriched fraction. In the phenobarbital-treated mice, the binding of both CAR and RXR was rapidly increased before the induction of CYP2B10 mRNA. In vitro-translated CAR bound to NR1, but only in the presence of similarly prepared RXR. PBREM was synergistically activated by transfection of CAR and RXR in HepG2 and HEK293 cells when the NR1 site was functional. A CAR-RXR heterodimer has thus been characterized as a trans-acting factor for the phenobarbital-inducible Cyp2b10 gene.
Figures
FIG. 1
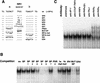
Functional assays to define the NR1 site as a DR4 motif. (A) The 51-bp enhancer–tk-CAT plasmids of the spacing and mutation variations of the 51-bp enhancer element were placed in front of the tk-CAT plasmids and were transfected into mouse primary hepatocytes. The CAT activity of the transfected cell extracts in the absence or the presence of 50 nM TCPOBOP is shown by open and solid bars, respectively. The data shown are means and standard deviations from three or four independent transfections, relative to the activity of the wild-type 51-bp enhancer element (wt = 100). (B) The PBREM sequence and motifs are depicted. The half-sites (a and b) of the putative NR binding sites are shown in boldface type. The bipartite NF1 binding motif is boxed.
FIG. 2
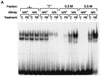
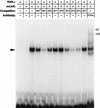
Gel shift assay to define the NR1 site as a DR4 motif and RXR binding. (A) Sequences of indicated NR1 mutants used as competitors for NR1 binding are compared to the wild-type NR1 (SP4 = wt). Spacer mutations are denoted by SP with numbers of bases. In MUT-SP, four bases of spacer are mutated. In addition to a random mutation in either the NR1a or NR1b site (NR1a mut and NR1b mut), these sites are mutated to create a perfect direct repeat with different orientations (PDR4, IR4, and ER4). The dots and hyphens indicate no change and deletions, respectively. The different nucleotides are shown in lowercase type. (B) Competition for NR1 binding was done with a 20-fold excess of indicated oligonucleotides. (C) Supershift assays were done by incubating preimmune IgG (1 μg) or indicated antibodies (1.5 μg) with liver nuclear extract from PB-treated mice. The results are representative of three independent experiments.
FIG. 3
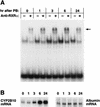
DNA affinity chromatography of RXRs and CAR. Pooled fractions of the liver nuclear extracts from a heparin-agarose column were incubated with either NR1 oligonucleotide-conjugated or NR1* oligonucleotide-conjugated magnetic beads. C and PB denote the extracts isolated from the untreated and the PB-treated (for 3 to 5 h) mice. The proteins were eluted from the beads with 0.3 and then 0.5 M NaCl. (A) The eluted proteins were subjected to a gel shift assay with NR1 as the probe. Nuclear extracts (1 μg of protein) and approximately 0.1 to 0.05 μg of the eluted proteins were used for gel shift assay. (B) For Western blots, 10 μg of nuclear extracts and 0.05 μg of the eluted proteins were resolved on an SDS–10% polyacrylamide gel and immunoblotted with anti-RXRα or anti-RXRβ IgG (1:3,000 dilution) or anti-hCAR serum (1:250 dilution). The arrows pointing to and from the bars indicate the applied and path-through fractions, respectively. Prestained Protein Marker Broad Range (New England Biolabs) was used as the molecular mass maker (shown in thousands). The results are representative of at least two independent purifications.
FIG. 3
DNA affinity chromatography of RXRs and CAR. Pooled fractions of the liver nuclear extracts from a heparin-agarose column were incubated with either NR1 oligonucleotide-conjugated or NR1* oligonucleotide-conjugated magnetic beads. C and PB denote the extracts isolated from the untreated and the PB-treated (for 3 to 5 h) mice. The proteins were eluted from the beads with 0.3 and then 0.5 M NaCl. (A) The eluted proteins were subjected to a gel shift assay with NR1 as the probe. Nuclear extracts (1 μg of protein) and approximately 0.1 to 0.05 μg of the eluted proteins were used for gel shift assay. (B) For Western blots, 10 μg of nuclear extracts and 0.05 μg of the eluted proteins were resolved on an SDS–10% polyacrylamide gel and immunoblotted with anti-RXRα or anti-RXRβ IgG (1:3,000 dilution) or anti-hCAR serum (1:250 dilution). The arrows pointing to and from the bars indicate the applied and path-through fractions, respectively. Prestained Protein Marker Broad Range (New England Biolabs) was used as the molecular mass maker (shown in thousands). The results are representative of at least two independent purifications.
FIG. 4
Supershift displaying the PB-dependent increase in RXR binding to NR1. (A) Liver nuclear extracts were prepared from 20 PB-treated mice injected at each time point. Three independent nuclear extracts were prepared from each time point, and 1 μg of the nuclear proteins per line was incubated with NR1 oligoprobe as described in Materials and Methods and resolved on a 5% polyacrylamide gel. The arrow indicates a band shifted by anti-RXRα. (B) Total RNA (6 μg per lane) prepared from the same liver pool at each time point was subjected to Northern hybridization with the CYP2B10 and albumin cDNA probes. The number above each lane indicates the time (in hours) after PB treatment. These results are representative of two or more independent experiments.
FIG. 5
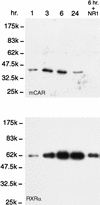
Western blot displaying the PB-dependent binding of mCAR1 and RXR to NR1. Liver nuclear extracts (1 mg of proteins in 1 ml of the incubation mixture) from the PB-treated mice at each time point (1 to 24 h) were incubated with NR1 affinity beads. In the right-hand lane (6 hr. + NR1), an excess amount of NR1 oligonucleotide (10 μg) was included as a competitor during the incubation. A 30-μl volume of the 100-μl eluates with 0.5 M NaCl was subjected to Western blot analysis with anti-RXRα IgG (1:3,000 dilution) and anti-CAR (peptide) serum (1:250 dilution). The results are representative of five or more independent experiments.
FIG. 6
Binding of in vitro-translated CAR and RXR to NR1 probe. Gel shifts were performed using in vitro-translated CARs (1.25 μl each) and RXR (1.25 μl) and 32P-labeled NR1 probe. For competitions, a 50-fold molar excess of each cold oligonucleotide was added to the reaction mixtures. The figure was generated from an experiment with mCAR since hCAR bound to the NR1 probe with the same specificity as did mCAR. The solid arrowhead indicates a gel shift band formed with RXR, mCAR, and NR1, while the open arrowheads point to supershift bands by anti-RXR. The results are representative of two or more independent experiments.
FIG. 7
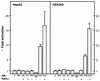
Activation of PBREM by CAR and RXR in the transformed cell lines. The HepG2 (or HEK293) cells were transfected at 30 to 40% confluence with 0.5 μg (or 0.4 μg) of the appropriate CAT reporter plasmid, 0.5 μg (or 0.125 μg) of β-galactosidase plasmid, and 0.15 μg (or 0.075 μg) of expression vector for individual nuclear receptors. At 48 h after transfection, cell extracts were assayed for β-galactosidase and CAT activities. The fold inductions of the normalized CAT activities are compared with the activity from the PBREM–tk-CAT alone (set to 100). Standard deviations were calculated from at least three independent experiments.
Similar articles
- The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene.
Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. Sueyoshi T, et al. J Biol Chem. 1999 Mar 5;274(10):6043-6. doi: 10.1074/jbc.274.10.6043. J Biol Chem. 1999. PMID: 10037683 - Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene.
Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Kawamoto T, et al. Mol Cell Biol. 1999 Sep;19(9):6318-22. doi: 10.1128/MCB.19.9.6318. Mol Cell Biol. 1999. PMID: 10454578 Free PMC article. - Phenobarbital induction of drug/steroid-metabolizing enzymes and nuclear receptor CAR.
Kakizaki S, Yamamoto Y, Ueda A, Moore R, Sueyoshi T, Negishi M. Kakizaki S, et al. Biochim Biophys Acta. 2003 Feb 17;1619(3):239-42. doi: 10.1016/s0304-4165(02)00482-8. Biochim Biophys Acta. 2003. PMID: 12573483 Review. - Phenobarbital response elements of cytochrome P450 genes and nuclear receptors.
Sueyoshi T, Negishi M. Sueyoshi T, et al. Annu Rev Pharmacol Toxicol. 2001;41:123-43. doi: 10.1146/annurev.pharmtox.41.1.123. Annu Rev Pharmacol Toxicol. 2001. PMID: 11264453 Review.
Cited by
- Complex effects of rexinoids on ligand dependent activation or inhibition of the xenobiotic receptor, CAR.
Tzameli I, Chua SS, Cheskis B, Moore DD. Tzameli I, et al. Nucl Recept. 2003 Jun 6;1(1):2. doi: 10.1186/1478-1336-1-2. Nucl Recept. 2003. PMID: 12904257 Free PMC article. - Towards a new age of virtual ADME/TOX and multidimensional drug discovery.
Ekins S, Boulanger B, Swaan PW, Hupcey MA. Ekins S, et al. Mol Divers. 2002;5(4):255-75. doi: 10.1023/a:1021376212320. Mol Divers. 2002. PMID: 12549676 Review. - Interactions that determine the assembly of a retinoid X receptor/corepressor complex.
Ghosh JC, Yang X, Zhang A, Lambert MH, Li H, Xu HE, Chen JD. Ghosh JC, et al. Proc Natl Acad Sci U S A. 2002 Apr 30;99(9):5842-7. doi: 10.1073/pnas.092043399. Epub 2002 Apr 23. Proc Natl Acad Sci U S A. 2002. PMID: 11972046 Free PMC article. - Guide to the Assessment of Mature Liver Gene Expression in Stem Cell-Derived Hepatocytes.
Zabulica M, Srinivasan RC, Vosough M, Hammarstedt C, Wu T, Gramignoli R, Ellis E, Kannisto K, Collin de l'Hortet A, Takeishi K, Soto-Gutierrez A, Strom SC. Zabulica M, et al. Stem Cells Dev. 2019 Jul 15;28(14):907-919. doi: 10.1089/scd.2019.0064. Stem Cells Dev. 2019. PMID: 31122128 Free PMC article. - The xenobiotic receptors PXR and CAR in liver physiology, an update.
Cai X, Young GM, Xie W. Cai X, et al. Biochim Biophys Acta Mol Basis Dis. 2021 Jun 1;1867(6):166101. doi: 10.1016/j.bbadis.2021.166101. Epub 2021 Feb 15. Biochim Biophys Acta Mol Basis Dis. 2021. PMID: 33600998 Free PMC article. Review.
References
- Choi H-S, Chung M, Tzameli I, Simha D, Lee Y-K, Seol W, Moore D D. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J Biol Chem. 1997;272:23565–23571. - PubMed
- Conney A H. Pharmacological implications of microsomal enzyme induction. Pharmacol Rev. 1967;19:317–366. - PubMed
- Denison M S, Whitlock J P., Jr Xenobiotic-inducible transcription of cytochrome P450 genes. J Biol Chem. 1995;270:18175–18178. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources