The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques - PubMed (original) (raw)
. 1998 Sep 21;188(6):1159-71.
doi: 10.1084/jem.188.6.1159.
M Halloran, D Schenten, J Lee, P Racz, K Tenner-Racz, J Manola, R Gelman, B Etemad-Moghadam, E Desjardins, R Wyatt, N P Gerard, L Marcon, D Margolin, J Fanton, M K Axthelm, N L Letvin, J Sodroski
Affiliations
- PMID: 9743534
- PMCID: PMC2212530
- DOI: 10.1084/jem.188.6.1159
The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques
G B Karlsson et al. J Exp Med. 1998.
Abstract
CD4+ T lymphocyte depletion in human immunodeficiency virus type 1 (HIV-1)-infected humans underlies the development of acquired immune deficiency syndrome. Using a model in which rhesus macaques were infected with chimeric simian-human immunodeficiency viruses (SHIVs), we show that both the level of viremia and the structure of the HIV-1 envelope glycoprotein ectodomains individually contributed to the efficiency with which CD4(+) T lymphocytes were depleted. The envelope glycoproteins of recombinant SHIVs that efficiently caused loss of CD4(+) T lymphocytes exhibited increased chemokine receptor binding and membrane-fusing capacity compared with those of less pathogenic viruses. These studies identify the HIV-1 envelope glycoprotein ectodomains as determinants of CD4(+) T lymphocyte loss in vivo and provide a foundation for studying pathogenic mechanisms.
Figures
Figure 1
Genomic organization of SHIV variants. The SHIV proviral genome is shown on the top. Sequences from SIVmac239 are shown in gray boxes and sequences from HIV-1 are shown in white (, 52). Enzyme restriction sites relevant for cloning of the SHIV variants are shown. The proviral structures of the SHIV variants used in this study are depicted beneath. SHIV-89.6* is identical to the parental SHIV-89.6, except that it contains the passage-associated tat and LTR changes (34). The other three variants differ from SHIV-89.6* only in the sequence of the envelope glycoproteins. SHIV-KB9 has the passage-associated amino acid changes in the gp120/gp41 ectodomains and the gp41 cytoplasmic tail change compared with SHIV-89.6*. SHIV-KB9ct has only the gp41 cytoplasmic tail change, and SHIV-KB9ecto has only the gp120/gp41 ectodomain changes.
Figure 2
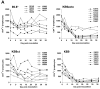
Absolute CD4+ T cell counts and plasma p27 antigenemia in infected monkeys. (A) The absolute CD4+ T cell counts in the peripheral blood of animals infected with SHIV-89.6*, SHIV-KB9ct, SHIV-KB9ecto, and SHIV-KB9 during the first 36 d of infection are shown. (B) The p27 antigen levels in the plasma of SHIV-infected animals are shown.
Figure 2
Absolute CD4+ T cell counts and plasma p27 antigenemia in infected monkeys. (A) The absolute CD4+ T cell counts in the peripheral blood of animals infected with SHIV-89.6*, SHIV-KB9ct, SHIV-KB9ecto, and SHIV-KB9 during the first 36 d of infection are shown. (B) The p27 antigen levels in the plasma of SHIV-infected animals are shown.
Figure 3
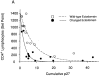
Relationship between viremia and CD4+ T lymphocyte counts for SHIV variants with different envelope glycoprotein domains. A “set-point” value for the absolute CD4+ T lymphocyte count in each animal (the median of the CD4 counts recorded on days 14–36 after infection) was plotted against the cumulative p27 antigenemia (area under the p27 versus time curves in Fig. 2_B_). The data were grouped according to the sequences (either wild-type 89.6 or containing the KB9 passage- associated changes) of the viral envelope glycoprotein ectodomains (A) or cytoplasmic tails (B). The curves are fitted to the data using a nonlinear exponential decay model.
Figure 3
Relationship between viremia and CD4+ T lymphocyte counts for SHIV variants with different envelope glycoprotein domains. A “set-point” value for the absolute CD4+ T lymphocyte count in each animal (the median of the CD4 counts recorded on days 14–36 after infection) was plotted against the cumulative p27 antigenemia (area under the p27 versus time curves in Fig. 2_B_). The data were grouped according to the sequences (either wild-type 89.6 or containing the KB9 passage- associated changes) of the viral envelope glycoprotein ectodomains (A) or cytoplasmic tails (B). The curves are fitted to the data using a nonlinear exponential decay model.
Figure 4
Relationship between infected lymph node cells and CD4+ T lymphocyte counts. The number of viral RNA–positive cells in the T cell zone of lymph nodes, which were taken from the animals at day 10 after inoculation, was compared with the “set point” values for the peripheral blood CD4+ T lymphocyte counts. The animal identification numbers for three of the data points discussed in the text are provided.
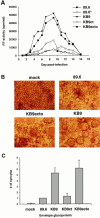
Figure 5
Replication and syncytium-forming ability of SHIV variants. (A) Rhesus PBMCs were PHA-stimulated and infected with equal amounts of SHIV-89.6, SHIV-89.6*, SHIV-KB9ct, SHIV-KB9ecto, and SHIV-KB9. Cells were maintained in the presence of IL-2 for the duration of the experiment and an aliquot of the culture medium was removed every day for RT analysis. (B) Uninfected CEM×174 cultures (mock) were compared with cultures infected with SHIV-89.6, SHIV-KB9ecto, and SHIV-KB9 for the presence of syncytia. (C) COS-1 cells, transiently expressing the 89.6, KB9ct, KB9ecto, or KB9 envelope glycoproteins, were cocultivated with CEM×174 cells for 6 h at 37°C. The number of syncytia was scored and normalized to that observed for the parental 89.6 envelope glycoproteins. The mean values and SE derived from three independent experiments are shown.
Figure 6
Receptor-binding of the 89.6 and KB9 gp120 envelope glycoproteins. (A) The binding of the 89.6 and KB9 gp120 glycoproteins to human sCD4 absorbed onto the surface of an ELISA plate is shown. Values represent the mean and standard deviations from two independent experiments, each containing four replicate samples. (B) The ability of the 89.6 and KB9 gp120 glycoproteins to compete with MIP-1β binding to mouse cells expressing human CCR5 was assessed in the presence of 100 nM sCD4. The results shown are representative of two independent experiments, each performed with duplicate samples at each concentration of competitor. Relative inhibitory constants for MIP-1β (positive control), 89.6 gp120, and KB9 gp120 were 0.20 nM, 39.0 nM, and 4.4 nM, respectively. The YU2ΔV1/2/3 glycoprotein, which lacks the V3 loop and thus is unable to bind CCR5 (19), was included as a negative control.
Figure 6
Receptor-binding of the 89.6 and KB9 gp120 envelope glycoproteins. (A) The binding of the 89.6 and KB9 gp120 glycoproteins to human sCD4 absorbed onto the surface of an ELISA plate is shown. Values represent the mean and standard deviations from two independent experiments, each containing four replicate samples. (B) The ability of the 89.6 and KB9 gp120 glycoproteins to compete with MIP-1β binding to mouse cells expressing human CCR5 was assessed in the presence of 100 nM sCD4. The results shown are representative of two independent experiments, each performed with duplicate samples at each concentration of competitor. Relative inhibitory constants for MIP-1β (positive control), 89.6 gp120, and KB9 gp120 were 0.20 nM, 39.0 nM, and 4.4 nM, respectively. The YU2ΔV1/2/3 glycoprotein, which lacks the V3 loop and thus is unable to bind CCR5 (19), was included as a negative control.
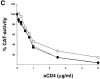
Figure 7
Neutralization of viruses with the 89.6, KB9ct and KB9 envelope glycoproteins. Recombinant viruses encoding CAT and bearing either the 89.6, KB9ct, or KB9 envelope glycoproteins were incubated with different concentrations of IgG1b12 (A), AG1121 (B), or sCD4 (C). The viruses were then incubated with CEM×174 cells. CAT activity in the CEM×174 cells was assessed 3 d later and is expressed as the percentage of CAT activity seen in the absence of antibody or sCD4. The results shown are representative of those obtained in at least two independent experiments.
Figure 7
Neutralization of viruses with the 89.6, KB9ct and KB9 envelope glycoproteins. Recombinant viruses encoding CAT and bearing either the 89.6, KB9ct, or KB9 envelope glycoproteins were incubated with different concentrations of IgG1b12 (A), AG1121 (B), or sCD4 (C). The viruses were then incubated with CEM×174 cells. CAT activity in the CEM×174 cells was assessed 3 d later and is expressed as the percentage of CAT activity seen in the absence of antibody or sCD4. The results shown are representative of those obtained in at least two independent experiments.
Similar articles
- Elite Control, Gut CD4 T Cell Sparing, and Enhanced Mucosal T Cell Responses in Macaca nemestrina Infected by a Simian Immunodeficiency Virus Lacking a gp41 Trafficking Motif.
Breed MW, Elser SE, Torben W, Jordan AP, Aye PP, Midkiff C, Schiro F, Sugimoto C, Alvarez-Hernandez X, Blair RV, Somasunderam A, Utay NS, Kuroda MJ, Pahar B, Wiseman RW, O'Connor DH, LaBranche CC, Montefiori DC, Marsh M, Li Y, Piatak M Jr, Lifson JD, Keele BF, Fultz PN, Lackner AA, Hoxie JA. Breed MW, et al. J Virol. 2015 Oct;89(20):10156-75. doi: 10.1128/JVI.01134-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223646 Free PMC article. - Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus.
Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK, Letvin NL, Sodroski J. Etemad-Moghadam B, et al. J Virol. 2001 Jun;75(12):5646-55. doi: 10.1128/JVI.75.12.5646-5655.2001. J Virol. 2001. PMID: 11356972 Free PMC article. - Differential Impact of In Vivo CD8+ T Lymphocyte Depletion in Controller versus Progressor Simian Immunodeficiency Virus-Infected Macaques.
Chowdhury A, Hayes TL, Bosinger SE, Lawson BO, Vanderford T, Schmitz JE, Paiardini M, Betts M, Chahroudi A, Estes JD, Silvestri G. Chowdhury A, et al. J Virol. 2015 Sep;89(17):8677-86. doi: 10.1128/JVI.00869-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063417 Free PMC article. - Understanding the basis of CD4(+) T-cell depletion in macaques infected by a simian-human immunodeficiency virus.
Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK, Letvin NL, Sodroski J. Etemad-Moghadam B, et al. Vaccine. 2002 May 6;20(15):1934-7. doi: 10.1016/s0264-410x(02)00072-5. Vaccine. 2002. PMID: 11983249 Review. - CD4-HIV-1 Envelope Interactions: Critical Insights for the Simian/HIV/Macaque Model.
Sharma A, Overbaugh J. Sharma A, et al. AIDS Res Hum Retroviruses. 2018 Sep;34(9):778-779. doi: 10.1089/AID.2018.0110. Epub 2018 Jul 9. AIDS Res Hum Retroviruses. 2018. PMID: 29886767 Free PMC article. Review.
Cited by
- Differential Pathogenicity of SHIV KB9 and 89.6 Env Correlates with Bystander Apoptosis Induction in CD4+ T cells.
Mehmetoglu-Gurbuz T, Joshi A, Garg H. Mehmetoglu-Gurbuz T, et al. Viruses. 2019 Oct 1;11(10):911. doi: 10.3390/v11100911. Viruses. 2019. PMID: 31581579 Free PMC article. - Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys.
Si Z, Cayabyab M, Sodroski J. Si Z, et al. J Virol. 2001 May;75(9):4208-18. doi: 10.1128/JVI.75.9.4208-4218.2001. J Virol. 2001. PMID: 11287570 Free PMC article. - High frequency of virus-specific B lymphocytes in germinal centers of simian-human immunodeficiency virus-infected rhesus monkeys.
Margolin DH, Saunders EF, Bronfin B, de Rosa N, Axthelm MK, Alvarez X, Letvin NL. Margolin DH, et al. J Virol. 2002 Apr;76(8):3965-73. doi: 10.1128/jvi.76.8.3965-3973.2002. J Virol. 2002. PMID: 11907236 Free PMC article. - HIV-1 escape from the CCR5 antagonist maraviroc associated with an altered and less-efficient mechanism of gp120-CCR5 engagement that attenuates macrophage tropism.
Roche M, Jakobsen MR, Sterjovski J, Ellett A, Posta F, Lee B, Jubb B, Westby M, Lewin SR, Ramsland PA, Churchill MJ, Gorry PR. Roche M, et al. J Virol. 2011 May;85(9):4330-42. doi: 10.1128/JVI.00106-11. Epub 2011 Feb 23. J Virol. 2011. PMID: 21345957 Free PMC article. - Genetic signatures of HIV-1 envelope-mediated bystander apoptosis.
Joshi A, Lee RT, Mohl J, Sedano M, Khong WX, Ng OT, Maurer-Stroh S, Garg H. Joshi A, et al. J Biol Chem. 2014 Jan 31;289(5):2497-514. doi: 10.1074/jbc.M113.514018. Epub 2013 Nov 21. J Biol Chem. 2014. PMID: 24265318 Free PMC article.
References
- Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. Isolation of a T lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. - PubMed
- Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. - PubMed
- Letvin NL. Animal models for the study of human immunodeficiency virus infections. Curr Opin Immunol. 1992;4:481–485. - PubMed
- Daar ES, Moudgil T, Meyer RD, Ho DD. Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324:961–964. - PubMed
- Clark SJ, Saag MS, Decker D, Campbell-Hill S, Robertson JL, Veldkamp PJ, Kappes JC, Hahn BH, Shaw GM. High titers of cytopathic virus in plasma of patients with symptomatic primary HIV-1 infection. N Engl J Med. 1991;324:954–960. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI028691/AI/NIAID NIH HHS/United States
- CA-50139/CA/NCI NIH HHS/United States
- R01 AI020729/AI/NIAID NIH HHS/United States
- R01 AI033832/AI/NIAID NIH HHS/United States
- K01 RR000163/RR/NCRR NIH HHS/United States
- AI-20729/AI/NIAID NIH HHS/United States
- R01 CA050139/CA/NCI NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- P51 RR000163/RR/NCRR NIH HHS/United States
- T32 RR007000/RR/NCRR NIH HHS/United States
- P51 RR000168/RR/NCRR NIH HHS/United States
- K26 RR000168/RR/NCRR NIH HHS/United States
- R37 AI020729/AI/NIAID NIH HHS/United States
- CA-06516/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials