Mitotic inactivation of a human SWI/SNF chromatin remodeling complex - PubMed (original) (raw)
Mitotic inactivation of a human SWI/SNF chromatin remodeling complex
S Sif et al. Genes Dev. 1998.
Abstract
During mitosis, chromatin is condensed into mitotic chromosomes and transcription is inhibited, processes that might be opposed by the chromatin remodeling activity of the SWI/SNF complexes. Brg1 and hBrm, which are components of human SWI/SNF (hSWI/SNF) complexes, were recently shown to be phosphorylated during mitosis. This suggested that phosphorylation might be used as a switch to modulate SWI/SNF activity. Using an epitope-tag strategy, we have purified hSWI/SNF complexes at different stages of the cell cycle, and found that hSWI/SNF was inactive in cells blocked in G2-M. Mitotic hSWI/SNF contained Brg1 but not hBrm, and was phosphorylated on at least two subunits, hSWI3 and Brg1. In vitro, active hSWI/SNF from asynchronous cells can be phosphorylated and inactivated by ERK1, and reactivated by dephosphorylation. hSWI/SNF isolated as cells traversed mitosis regained activity when its subunits were dephosphorylated either in vitro or in vivo. We propose that this transitional inactivation and reactivation of hSWI/SNF is required for formation of a repressed chromatin structure during mitosis and reformation of an active chromatin structure as cells leave mitosis.
Figures
Figure 1
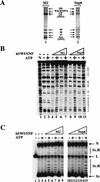
Purification of proteins associated with Flag-tagged Ini1. (A) Purification of Flag-tagged hSWI/SNF complexes. FL-Ini1-11 nuclear extracts were incubated with anti-Flag M2 affinity gel, and after several washes with buffer containing increasing amounts of salt, the proteins retained on the affinity column were eluted with buffer containing 20-fold molar excess of Flag peptide. Eluted proteins were then fractionated on a Superose 6 sizing column. (B) hSWI/SNF subunits cofractionate with FL-Ini1. Western blot analysis was performed with 50 μg of nuclear extract (NE); 1.2 μg of affinity purified SWI/SNF fraction (M2), and 15 μl of sizing column fractions 14–23 [Superose 6 (Sup6)].
Figure 1
Purification of proteins associated with Flag-tagged Ini1. (A) Purification of Flag-tagged hSWI/SNF complexes. FL-Ini1-11 nuclear extracts were incubated with anti-Flag M2 affinity gel, and after several washes with buffer containing increasing amounts of salt, the proteins retained on the affinity column were eluted with buffer containing 20-fold molar excess of Flag peptide. Eluted proteins were then fractionated on a Superose 6 sizing column. (B) hSWI/SNF subunits cofractionate with FL-Ini1. Western blot analysis was performed with 50 μg of nuclear extract (NE); 1.2 μg of affinity purified SWI/SNF fraction (M2), and 15 μl of sizing column fractions 14–23 [Superose 6 (Sup6)].
Figure 2
Biochemical characterization of Flag-tagged hSWI/SNF complexes. (A) Amounts of 560 ng of affinity-purified hSWI/SNF (M2, lane 1) and 465 ng of sizing column fraction Superose 6 (sup6), lane _2_] were analyzed by SDS-PAGE, and proteins were visualized by silver staining. (B) Nucleosome disrupting activity. Either 280 ng (lanes 4,5) or 560 ng (lanes 6,7) of M2 fractions was incubated with mononucleosomes with or without ATP. Similarly, 155 ng (lanes 8,9) and 310 ng (lanes 10,11) of Sup6 peak fractions were incubated with core particles as indicated. As a control, naked DNA (N, lane 1) and nucleosomal DNA with or without ATP (lanes 2,3) are shown. Symbols at right indicate the changes to DNase sensitivity caused by hSWI/SNF in an ATP-dependent manner. (C) Remodeling activity on nucleosomal arrays. Increasing amounts of either M2: 19 ng (lanes 4,5), 56 ng (lanes 6,7), 84 ng (lanes 8,9), or Sup6 fractions: 10 ng (lanes 10,11), 31 ng (lanes 12,13), 62 ng (lanes 14,15), were incubated with 8 ng of assembled chromatin templates with or without ATP as indicated. (Lane 1) Linear plasmid DNA (lanes 2,3) assembled template incubated with or without ATP. The resolved DNA templates are supercoiled (Sc); relaxed and supercoiled (Sc.R); linear (L); and nicked (N).
Figure 3
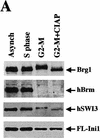
Mitotic hSWI/SNF is phosphorylated and lacks nucleosome disrupting activity. (A) hSWI/SNF subunits are regulated differently during mitosis. Approximately 10 μg of total protein from exponentially growing cells (Asynch), cells blocked in S phase (S phase), or cells blocked in prometaphase (G2–M) were analyzed by Western blotting with anti-Brg1, anti-hBrm, anti-hSWI3, or anti-Flag antiserum. Where indicated, proteins were incubated with 10–20 units of calf alkaline phosphatase (CIAP) for 45 min at 37°C. (B) Mitotic hSWI/SNF subunits cofractionate through Superose 6 (Sup6) sizing column. Western blot analysis of Sup6 column fractions (fr.) 15–27, with anti-Brg1, anti-hSWI3, and anti-Flag antisera. Mitotic hSWI/SNF subunits consistently (n = 4) eluted later (fr. 23) than the asynchronous complex (fr. 19). (C) Mitotic hSWI/SNF complex is intact. Equal amounts of hSWI/SNF sizing column peak fractions (∼500 ng) from either exponentially growing cells (Asynch, fr. 19), or cells blocked in early mitosis (G2–M, fr. 23) were analyzed by SDS-PAGE and silver staining. (D) Phosphatase treatment of mitotic hSWI/SNF enables the complex to disrupt mononucleosomes. Similar amounts of Sup6 fractions from either asynchronous cells (500 ng) or cells blocked in G2-M (560 ng) were incubated with mononucleosomes with or without ATP and CIAP as indicated.
Figure 3
Mitotic hSWI/SNF is phosphorylated and lacks nucleosome disrupting activity. (A) hSWI/SNF subunits are regulated differently during mitosis. Approximately 10 μg of total protein from exponentially growing cells (Asynch), cells blocked in S phase (S phase), or cells blocked in prometaphase (G2–M) were analyzed by Western blotting with anti-Brg1, anti-hBrm, anti-hSWI3, or anti-Flag antiserum. Where indicated, proteins were incubated with 10–20 units of calf alkaline phosphatase (CIAP) for 45 min at 37°C. (B) Mitotic hSWI/SNF subunits cofractionate through Superose 6 (Sup6) sizing column. Western blot analysis of Sup6 column fractions (fr.) 15–27, with anti-Brg1, anti-hSWI3, and anti-Flag antisera. Mitotic hSWI/SNF subunits consistently (n = 4) eluted later (fr. 23) than the asynchronous complex (fr. 19). (C) Mitotic hSWI/SNF complex is intact. Equal amounts of hSWI/SNF sizing column peak fractions (∼500 ng) from either exponentially growing cells (Asynch, fr. 19), or cells blocked in early mitosis (G2–M, fr. 23) were analyzed by SDS-PAGE and silver staining. (D) Phosphatase treatment of mitotic hSWI/SNF enables the complex to disrupt mononucleosomes. Similar amounts of Sup6 fractions from either asynchronous cells (500 ng) or cells blocked in G2-M (560 ng) were incubated with mononucleosomes with or without ATP and CIAP as indicated.
Figure 3
Mitotic hSWI/SNF is phosphorylated and lacks nucleosome disrupting activity. (A) hSWI/SNF subunits are regulated differently during mitosis. Approximately 10 μg of total protein from exponentially growing cells (Asynch), cells blocked in S phase (S phase), or cells blocked in prometaphase (G2–M) were analyzed by Western blotting with anti-Brg1, anti-hBrm, anti-hSWI3, or anti-Flag antiserum. Where indicated, proteins were incubated with 10–20 units of calf alkaline phosphatase (CIAP) for 45 min at 37°C. (B) Mitotic hSWI/SNF subunits cofractionate through Superose 6 (Sup6) sizing column. Western blot analysis of Sup6 column fractions (fr.) 15–27, with anti-Brg1, anti-hSWI3, and anti-Flag antisera. Mitotic hSWI/SNF subunits consistently (n = 4) eluted later (fr. 23) than the asynchronous complex (fr. 19). (C) Mitotic hSWI/SNF complex is intact. Equal amounts of hSWI/SNF sizing column peak fractions (∼500 ng) from either exponentially growing cells (Asynch, fr. 19), or cells blocked in early mitosis (G2–M, fr. 23) were analyzed by SDS-PAGE and silver staining. (D) Phosphatase treatment of mitotic hSWI/SNF enables the complex to disrupt mononucleosomes. Similar amounts of Sup6 fractions from either asynchronous cells (500 ng) or cells blocked in G2-M (560 ng) were incubated with mononucleosomes with or without ATP and CIAP as indicated.
Figure 3
Mitotic hSWI/SNF is phosphorylated and lacks nucleosome disrupting activity. (A) hSWI/SNF subunits are regulated differently during mitosis. Approximately 10 μg of total protein from exponentially growing cells (Asynch), cells blocked in S phase (S phase), or cells blocked in prometaphase (G2–M) were analyzed by Western blotting with anti-Brg1, anti-hBrm, anti-hSWI3, or anti-Flag antiserum. Where indicated, proteins were incubated with 10–20 units of calf alkaline phosphatase (CIAP) for 45 min at 37°C. (B) Mitotic hSWI/SNF subunits cofractionate through Superose 6 (Sup6) sizing column. Western blot analysis of Sup6 column fractions (fr.) 15–27, with anti-Brg1, anti-hSWI3, and anti-Flag antisera. Mitotic hSWI/SNF subunits consistently (n = 4) eluted later (fr. 23) than the asynchronous complex (fr. 19). (C) Mitotic hSWI/SNF complex is intact. Equal amounts of hSWI/SNF sizing column peak fractions (∼500 ng) from either exponentially growing cells (Asynch, fr. 19), or cells blocked in early mitosis (G2–M, fr. 23) were analyzed by SDS-PAGE and silver staining. (D) Phosphatase treatment of mitotic hSWI/SNF enables the complex to disrupt mononucleosomes. Similar amounts of Sup6 fractions from either asynchronous cells (500 ng) or cells blocked in G2-M (560 ng) were incubated with mononucleosomes with or without ATP and CIAP as indicated.
Figure 4
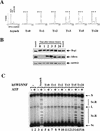
Cell cycle-dependent reactivation of hSWI/SNF complexes. (A) Exponentially growing FL-Ini1 cells were synchronized in G2–M by nocodazole treatment, and then released from the block by washing in medium without drug. Cells were harvested at the indicated times (T), and their DNA content was determined by FACS analysis. For comparison, the DNA content of exponentially growing cells (Asynch) is shown. Small arrowheads indicate the position of each stage of the cell cycle. (B) Cell cycle-dependent dephosphorylation of hSWI/SNF subunits. hSWI/SNF complexes were affinity purified from either asynchronous cells (Asynch), or cells blocked in G2–M and then released (T = 0, 1, 2, 3, 5, and 24), and hSWI/SNF fractions were then examined for their protein composition and phosphorylation state. Approximately 200 ng of each fraction was analyzed by Western blotting with either anti-Brg1, anti-hBrm, or anti-hSWI3. (C) hSWI/SNF reactivation correlates with dephosphorylation of its subunits. Equal amounts (200 ng) of affinity-purified hSWI/SNF fractions from asynchronous cells (lanes 3,4), cells blocked in G2–M (lanes 5,6), or cells that were released from the block (lanes 7–16) were tested for chromatin remodeling activity by use of an 8 ng assembled template, with or without ATP as indicated. Nicked closed circular (N); linear (L); supercoiled (Sc); and supercoiled and relaxed (Sc.R) plasmid DNA are indicated.
Figure 5
Active hSWI/SNF is inhibited by ERK1 in vitro. (A) Brg1, hBrm, and hSWI3 are phosphorylated in vitro by GST–ERK1. Equal amounts (0.75 μg/15 μl reaction) of either wild-type GST–ERK1 or mutant GST–ERK1 (K63M) were activated by GST–His–MEK1 (0.25 μg/15 μl reaction) as described in Materials and Methods. Approximately 200 ng of asynchronous SWI/SNF was added to the reactions containing either GST–ERK1 (lane 2), or GST–ERK1 (K63M) (lane 3). Samples were incubated at 30°C for 1 hr and analyzed by Western blotting. When PP2A was added (lane 4), the GST fusion proteins were removed by adding GST beads to the reactions, and the supernatant was incubated with 0.1 units of PP2A at 30°C for 1 hr. As a control, asynchronous hSWI/SNF subunits are shown (lanes 1,5). (B) hSWI/SNF that is phosphorylated in vitro by ERK1 is inactive. Asynchronous hSWI/SNF fractions were incubated (conditions as in A) with either MEK1-activated or inactive GST–ERK1 and GST–ERK1 (K63M) as indicated. As controls, hSWI/SNF was also tested for activity after incubation with either GST–MEK1 (lane 5), GST–ERK1 (lane 6), or GST–ERK1 (K63M) (lane 7). When PP2A was added (lanes 10–15), samples were treated as described in A. Assays are performed as in Fig. 4. (C) Brg1-containing complex is efficiently reactivated by PP2A. hSWI/SNF fractions purified from cells growing exponentially (Asynch), cells blocked in G2–M (T = 0), or cells blocked and then released (T = 1), were incubated with supercoiled templates with or without ATP and PP2A as indicated. When PP2A was added, samples were preincubated with 0.1 units of PP2A at 30°C for 1 hr.
Figure 5
Active hSWI/SNF is inhibited by ERK1 in vitro. (A) Brg1, hBrm, and hSWI3 are phosphorylated in vitro by GST–ERK1. Equal amounts (0.75 μg/15 μl reaction) of either wild-type GST–ERK1 or mutant GST–ERK1 (K63M) were activated by GST–His–MEK1 (0.25 μg/15 μl reaction) as described in Materials and Methods. Approximately 200 ng of asynchronous SWI/SNF was added to the reactions containing either GST–ERK1 (lane 2), or GST–ERK1 (K63M) (lane 3). Samples were incubated at 30°C for 1 hr and analyzed by Western blotting. When PP2A was added (lane 4), the GST fusion proteins were removed by adding GST beads to the reactions, and the supernatant was incubated with 0.1 units of PP2A at 30°C for 1 hr. As a control, asynchronous hSWI/SNF subunits are shown (lanes 1,5). (B) hSWI/SNF that is phosphorylated in vitro by ERK1 is inactive. Asynchronous hSWI/SNF fractions were incubated (conditions as in A) with either MEK1-activated or inactive GST–ERK1 and GST–ERK1 (K63M) as indicated. As controls, hSWI/SNF was also tested for activity after incubation with either GST–MEK1 (lane 5), GST–ERK1 (lane 6), or GST–ERK1 (K63M) (lane 7). When PP2A was added (lanes 10–15), samples were treated as described in A. Assays are performed as in Fig. 4. (C) Brg1-containing complex is efficiently reactivated by PP2A. hSWI/SNF fractions purified from cells growing exponentially (Asynch), cells blocked in G2–M (T = 0), or cells blocked and then released (T = 1), were incubated with supercoiled templates with or without ATP and PP2A as indicated. When PP2A was added, samples were preincubated with 0.1 units of PP2A at 30°C for 1 hr.
Figure 5
Active hSWI/SNF is inhibited by ERK1 in vitro. (A) Brg1, hBrm, and hSWI3 are phosphorylated in vitro by GST–ERK1. Equal amounts (0.75 μg/15 μl reaction) of either wild-type GST–ERK1 or mutant GST–ERK1 (K63M) were activated by GST–His–MEK1 (0.25 μg/15 μl reaction) as described in Materials and Methods. Approximately 200 ng of asynchronous SWI/SNF was added to the reactions containing either GST–ERK1 (lane 2), or GST–ERK1 (K63M) (lane 3). Samples were incubated at 30°C for 1 hr and analyzed by Western blotting. When PP2A was added (lane 4), the GST fusion proteins were removed by adding GST beads to the reactions, and the supernatant was incubated with 0.1 units of PP2A at 30°C for 1 hr. As a control, asynchronous hSWI/SNF subunits are shown (lanes 1,5). (B) hSWI/SNF that is phosphorylated in vitro by ERK1 is inactive. Asynchronous hSWI/SNF fractions were incubated (conditions as in A) with either MEK1-activated or inactive GST–ERK1 and GST–ERK1 (K63M) as indicated. As controls, hSWI/SNF was also tested for activity after incubation with either GST–MEK1 (lane 5), GST–ERK1 (lane 6), or GST–ERK1 (K63M) (lane 7). When PP2A was added (lanes 10–15), samples were treated as described in A. Assays are performed as in Fig. 4. (C) Brg1-containing complex is efficiently reactivated by PP2A. hSWI/SNF fractions purified from cells growing exponentially (Asynch), cells blocked in G2–M (T = 0), or cells blocked and then released (T = 1), were incubated with supercoiled templates with or without ATP and PP2A as indicated. When PP2A was added, samples were preincubated with 0.1 units of PP2A at 30°C for 1 hr.
Similar articles
- Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits.
Phelan ML, Sif S, Narlikar GJ, Kingston RE. Phelan ML, et al. Mol Cell. 1999 Feb;3(2):247-53. doi: 10.1016/s1097-2765(00)80315-9. Mol Cell. 1999. PMID: 10078207 - Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes.
Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Sif S, et al. Genes Dev. 2001 Mar 1;15(5):603-18. doi: 10.1101/gad.872801. Genes Dev. 2001. PMID: 11238380 Free PMC article. - Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases.
Phelan ML, Schnitzler GR, Kingston RE. Phelan ML, et al. Mol Cell Biol. 2000 Sep;20(17):6380-9. doi: 10.1128/MCB.20.17.6380-6389.2000. Mol Cell Biol. 2000. PMID: 10938115 Free PMC article. - SWI/SNF complex: dissection of a chromatin remodeling cycle.
Peterson CL. Peterson CL. Cold Spring Harb Symp Quant Biol. 1998;63:545-52. doi: 10.1101/sqb.1998.63.545. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384319 Review. No abstract available. - When the SWI/SNF complex remodels...the cell cycle.
Muchardt C, Yaniv M. Muchardt C, et al. Oncogene. 2001 May 28;20(24):3067-75. doi: 10.1038/sj.onc.1204331. Oncogene. 2001. PMID: 11420722 Review.
Cited by
- Cooperativity of imprinted genes inactivated by acquired chromosome 20q deletions.
Aziz A, Baxter EJ, Edwards C, Cheong CY, Ito M, Bench A, Kelley R, Silber Y, Beer PA, Chng K, Renfree MB, McEwen K, Gray D, Nangalia J, Mufti GJ, Hellstrom-Lindberg E, Kiladjian JJ, McMullin MF, Campbell PJ, Ferguson-Smith AC, Green AR. Aziz A, et al. J Clin Invest. 2013 May;123(5):2169-82. doi: 10.1172/JCI66113. Epub 2013 Apr 1. J Clin Invest. 2013. PMID: 23543057 Free PMC article. - MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation.
Stojanova A, Tu WB, Ponzielli R, Kotlyar M, Chan PK, Boutros PC, Khosravi F, Jurisica I, Raught B, Penn LZ. Stojanova A, et al. Cell Cycle. 2016 Jul 2;15(13):1693-705. doi: 10.1080/15384101.2016.1146836. Epub 2016 Jun 7. Cell Cycle. 2016. PMID: 27267444 Free PMC article. - Nuclear localization signal region in nuclear receptor PXR governs the receptor association with mitotic chromatin.
Rana M, Dash AK, Ponnusamy K, Tyagi RK. Rana M, et al. Chromosome Res. 2018 Dec;26(4):255-276. doi: 10.1007/s10577-018-9583-2. Epub 2018 Jul 15. Chromosome Res. 2018. PMID: 30009337 - SNR1 (INI1/SNF5) mediates important cell growth functions of the Drosophila Brahma (SWI/SNF) chromatin remodeling complex.
Zraly CB, Marenda DR, Dingwall AK. Zraly CB, et al. Genetics. 2004 Sep;168(1):199-214. doi: 10.1534/genetics.104.029439. Genetics. 2004. PMID: 15454538 Free PMC article. - Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes.
Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Pal S, et al. Mol Cell Biol. 2004 Nov;24(21):9630-45. doi: 10.1128/MCB.24.21.9630-9645.2004. Mol Cell Biol. 2004. PMID: 15485929 Free PMC article.
References
- Acharya U, Mallabiabarrena A, Acharya JK, Malhotra V. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for golgi fragmentation during mitosis. Cell. 1998;92:183–192. - PubMed
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley and Sons; 1996.
- Bulger M, Kadonaga JT. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;14:241–262.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous