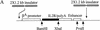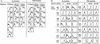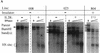Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators - PubMed (original) (raw)
Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators
M J Pikaart et al. Genes Dev. 1998.
Abstract
The constitutive DNase I hypersensitive site at the 5' end of the chicken beta-globin locus marks the boundary of the active chromatin domain in erythroid cells. The DNA sequence containing this site has the properties of an insulator, as shown by its ability in stable transformation experiments to block enhancer-promoter interaction when it lies between the two, but not when it lies outside, and to protect against position effects in Drosophila. We now show that the chicken insulator can protect a stably integrated gene, which is otherwise subject to great variability of expression, from chromatin-mediated repression in cell culture. When the integrated reporter gene is surrounded by insulator elements, stably transformed cell lines display consistent enhancer-dependent expression levels, in accord with the strength of the enhancer. In the absence of insulators, long-term nonselective propagation of cells carrying the integrated reporter gene results in gradual extinction of the reporter's expression, with expression patterns from tandemly repeated inserted genes suggesting that the extinction of adjacent genes is coupled. We show that the uninsulated reporter genes, in addition to becoming transcriptionally inactive, lose several epigenetic hallmarks of active chromatin, including nuclease accessibility, DNA hypomethylation, and histone hyperacetylation during time in culture. Treatment with inhibitors of histone deacetylase or DNA methylation reverses the extinction of the uninsulated genes. Extinction is completely prevented by flanking the reporter construct with insulators. Furthermore, in contrast to the uninsulated reporter genes, chromatin over the insulated genes retains nuclease accessibility and histone hyperacetylation. However, there is no clear correlation between the presence of the insulators and the level of DNA methylation. This leads us to propose a model for the insulator's ability to protect against extinction in the transformed cell lines and to function as a chromatin boundary for the chicken beta-globin locus in normal erythroid cells.
Figures
Figure 1
(A) The IL2R cDNA and SV40 splice/polyadenylation signal are linked to the chicken βA-globin promoter and a 121-bp minimal β/ε enhancer element or enhancers mutated at either the NF–E2 site or at both GATA sites. These mutations decrease expression to 10% and 6%, respectively, of that of the wild-type (Reitman and Felsenfeld 1988) and reduce the probability of forming a hypersensitive site over the enhancer when the construct is stably integrated in 6C2 cells (Boyes and Felsenfeld 1996). Furthermore, the expression cassette was flanked with two copies of the 1.2-kb globin insulator. (B) FACS profiles of 6C2 cells untransfected with the IL2R construct (left) and following stable transfection and integration of one copy of the IL2R reporter with wild-type enhancer (right; these are line 005 cells, below). On these histograms, cell number is indicated on the y-axis, and the logarithm (10−1 through 103) of FITC fluorescence intensity on the x-axis. The horizontal bars denoted B and C define the ranges of IL2R negative and positive cells, respectively.
Figure 1
(A) The IL2R cDNA and SV40 splice/polyadenylation signal are linked to the chicken βA-globin promoter and a 121-bp minimal β/ε enhancer element or enhancers mutated at either the NF–E2 site or at both GATA sites. These mutations decrease expression to 10% and 6%, respectively, of that of the wild-type (Reitman and Felsenfeld 1988) and reduce the probability of forming a hypersensitive site over the enhancer when the construct is stably integrated in 6C2 cells (Boyes and Felsenfeld 1996). Furthermore, the expression cassette was flanked with two copies of the 1.2-kb globin insulator. (B) FACS profiles of 6C2 cells untransfected with the IL2R construct (left) and following stable transfection and integration of one copy of the IL2R reporter with wild-type enhancer (right; these are line 005 cells, below). On these histograms, cell number is indicated on the y-axis, and the logarithm (10−1 through 103) of FITC fluorescence intensity on the x-axis. The horizontal bars denoted B and C define the ranges of IL2R negative and positive cells, respectively.
Figure 2
FACS analyses for position effects (A) Cell lines carrying single copies of the IL2R constructs with or without flanking insulators were grown under selection for the hygromycin resistance marker and analyzed by FACS for IL2R cell surface activity. The most left-shifted histogram peaks, such as line 707, fall at the fluorescence intensity position of control, untransfected 6C2 cells analyzed under the same conditions (see Fig. 1B). (B) Wild-type enhancer lines, in single- and multiple-copy, both with and without insulators, were grown for up to 100 days in culture without hygromycin. Cells were analyzed during the time course for IL2R activity. Of the uninsulated lines tested, only one (005) remained active, whereas all lines with insulators retained full activity throughout passage. The rightward peak shifts of lines 013, 025, 809, and 804, reflect increased IL2R mRNA levels caused by multiple gene copies. When RNA was prepared from sorted cells, IL2R message levels were found to be unchanged in the active fractions relative to the initial, 100% active state, whereas message was undetectable in the inactive fractions (data not shown).
Figure 3
Nuclease accessibility in IL2R active and inactive cells. (A) A single-copy (008) and a multiple-copy (025) cell line at ∼50 days following release from hygromycin selection were sorted by MACS into IL2R inactive (−) and active (+) fractions. Nuclei from the sorted cells were digested with increasing concentrations of DNase I, as were nuclei from line 804, which carries multiple copies of the insulated construct and is fully active. DNA samples were analyzed by digesting with _Bam_HI, electrophoresis and Southern blotting, and probing by hybridization to a radiolabeled DNA corresponding to the coding sequence of the IL2R cDNA (_Bam_HI–_Xba_I fragment—Fig. 1). The arrays of parent _Bam_HI bands in lines 025 and 804 are indicative of multiple integrants. Line 008 has one _Bam_HI parent band at single-copy intensity; the second band below it hybridizes to probe at less than single-copy intensity, and on the basis of the sizes of several restriction digestion products, appears to be a separately integrated fragment of the IL2R construct that has lost the promoter and 3′ portion of the gene. (B) Single- and multiple-copy uninsulated lines were sorted by MACS as in A and nuclei were digested with _Pvu_II, as were nuclei from unsorted, active single and multiple-copy insulated lines. _Pvu_II produces a doublet of digested bands (asterisks) as a result of the two adjacent sites, one inside the enhancer fragment and one just outside it. Percent digestion was calculated by measuring band intensities on a PhophorImager and taking the ratio of the sum of the digested band doublet to the total of the parent (arrow) and digested bands. As a control for the extent of nuclease digestion, blots were stripped of labeled IL2R probe and reprobed with sequence specific to the chicken βA gene-coding region (not in the IL2R reporter plasmids) such that cutting was detected at the endogenous β/ε enhancer, whose accessibility to DNase I and _Pvu_II was equivalent for all lines examined (data not shown). For most cell lines, sorting and nuclear isolation followed by DNase I or _Pvu_II digestion was repeated two or three times. When repeated, the extent of digestion by the two nucleases was always reproducible.
Figure 3
Nuclease accessibility in IL2R active and inactive cells. (A) A single-copy (008) and a multiple-copy (025) cell line at ∼50 days following release from hygromycin selection were sorted by MACS into IL2R inactive (−) and active (+) fractions. Nuclei from the sorted cells were digested with increasing concentrations of DNase I, as were nuclei from line 804, which carries multiple copies of the insulated construct and is fully active. DNA samples were analyzed by digesting with _Bam_HI, electrophoresis and Southern blotting, and probing by hybridization to a radiolabeled DNA corresponding to the coding sequence of the IL2R cDNA (_Bam_HI–_Xba_I fragment—Fig. 1). The arrays of parent _Bam_HI bands in lines 025 and 804 are indicative of multiple integrants. Line 008 has one _Bam_HI parent band at single-copy intensity; the second band below it hybridizes to probe at less than single-copy intensity, and on the basis of the sizes of several restriction digestion products, appears to be a separately integrated fragment of the IL2R construct that has lost the promoter and 3′ portion of the gene. (B) Single- and multiple-copy uninsulated lines were sorted by MACS as in A and nuclei were digested with _Pvu_II, as were nuclei from unsorted, active single and multiple-copy insulated lines. _Pvu_II produces a doublet of digested bands (asterisks) as a result of the two adjacent sites, one inside the enhancer fragment and one just outside it. Percent digestion was calculated by measuring band intensities on a PhophorImager and taking the ratio of the sum of the digested band doublet to the total of the parent (arrow) and digested bands. As a control for the extent of nuclease digestion, blots were stripped of labeled IL2R probe and reprobed with sequence specific to the chicken βA gene-coding region (not in the IL2R reporter plasmids) such that cutting was detected at the endogenous β/ε enhancer, whose accessibility to DNase I and _Pvu_II was equivalent for all lines examined (data not shown). For most cell lines, sorting and nuclear isolation followed by DNase I or _Pvu_II digestion was repeated two or three times. When repeated, the extent of digestion by the two nucleases was always reproducible.
Figure 4
The effects of inhibitors of histone deacetylation and DNA methylation on transcription. (A) Histone deacetylase inhibitors reverse extinction of expression. Cells were sorted with magnetic beads, and the inactive fractions were replated and grown in either untreated medium for 3 days (top), or in medium plus 5 ng/ml TSA and 50 m
m
Na butyrate for 24 hr followed by washing and 2 days of recovery in untreated medium (bottom). On the third day, cells were analyzed by FACS for IL2R expression. The vertical line in each FACS panel marks the cutoff observed between inactive and active cells, and the number in the upper right corner of each indicates the percentage of total cells that fluoresce at intensities higher than that cutoff. (B) Inactive cells were treated with butyrate or TSA separately (same conditions as in A). (C) Inactive cells grown for 2 days untreated or in the presence of 50 μ
m
5-azaC, then assayed for IL2R by FACS. Percentages of cells in the active fluorescent range are denoted as in A.
Figure 4
The effects of inhibitors of histone deacetylation and DNA methylation on transcription. (A) Histone deacetylase inhibitors reverse extinction of expression. Cells were sorted with magnetic beads, and the inactive fractions were replated and grown in either untreated medium for 3 days (top), or in medium plus 5 ng/ml TSA and 50 m
m
Na butyrate for 24 hr followed by washing and 2 days of recovery in untreated medium (bottom). On the third day, cells were analyzed by FACS for IL2R expression. The vertical line in each FACS panel marks the cutoff observed between inactive and active cells, and the number in the upper right corner of each indicates the percentage of total cells that fluoresce at intensities higher than that cutoff. (B) Inactive cells were treated with butyrate or TSA separately (same conditions as in A). (C) Inactive cells grown for 2 days untreated or in the presence of 50 μ
m
5-azaC, then assayed for IL2R by FACS. Percentages of cells in the active fluorescent range are denoted as in A.
Figure 4
The effects of inhibitors of histone deacetylation and DNA methylation on transcription. (A) Histone deacetylase inhibitors reverse extinction of expression. Cells were sorted with magnetic beads, and the inactive fractions were replated and grown in either untreated medium for 3 days (top), or in medium plus 5 ng/ml TSA and 50 m
m
Na butyrate for 24 hr followed by washing and 2 days of recovery in untreated medium (bottom). On the third day, cells were analyzed by FACS for IL2R expression. The vertical line in each FACS panel marks the cutoff observed between inactive and active cells, and the number in the upper right corner of each indicates the percentage of total cells that fluoresce at intensities higher than that cutoff. (B) Inactive cells were treated with butyrate or TSA separately (same conditions as in A). (C) Inactive cells grown for 2 days untreated or in the presence of 50 μ
m
5-azaC, then assayed for IL2R by FACS. Percentages of cells in the active fluorescent range are denoted as in A.
Figure 5
Histone acetylation assay on the uninsulated lines 013 and 025 and insulated line 804. Chromatin was immunoprecipitated with either no primary antibody (Con) as a control or with monoclonal antibody directed against acetylated histone H3 or H4 tails. (A) DNA from immunoprecipitated (IP) or free (F) histone complexes was subjected to PCR with primers specific to IL2R and globin sequences. The abundance of IL2R DNA detected in the fractions relative to the globin internal control was determined for three separate PCR reactions and given as an average below each lane, with standard deviations of 20% or less for each. IL2R to globin ratios between 4 and 6 are characteristic of genomic DNA; ratios substantially higher than this indicate relative hyperacetylation of histones bound to the IL2R sequence. (B) To examine the acetylation level of histones bound to transfected DNA outside the insulator sequences, the same immunoprecipitated DNA samples were amplified with a pair of primers directed to vector sequences and the globin control primers. All lines were assayed before release of hygromycin selection (top), when 013, 025, and 804 are all fully active, and after 80 days of nonselective growth (bottom), after which lines 013, 025 were nearly completely inactive while line 804 retained activity (see Fig. 2B).
Figure 5
Histone acetylation assay on the uninsulated lines 013 and 025 and insulated line 804. Chromatin was immunoprecipitated with either no primary antibody (Con) as a control or with monoclonal antibody directed against acetylated histone H3 or H4 tails. (A) DNA from immunoprecipitated (IP) or free (F) histone complexes was subjected to PCR with primers specific to IL2R and globin sequences. The abundance of IL2R DNA detected in the fractions relative to the globin internal control was determined for three separate PCR reactions and given as an average below each lane, with standard deviations of 20% or less for each. IL2R to globin ratios between 4 and 6 are characteristic of genomic DNA; ratios substantially higher than this indicate relative hyperacetylation of histones bound to the IL2R sequence. (B) To examine the acetylation level of histones bound to transfected DNA outside the insulator sequences, the same immunoprecipitated DNA samples were amplified with a pair of primers directed to vector sequences and the globin control primers. All lines were assayed before release of hygromycin selection (top), when 013, 025, and 804 are all fully active, and after 80 days of nonselective growth (bottom), after which lines 013, 025 were nearly completely inactive while line 804 retained activity (see Fig. 2B).
Figure 6
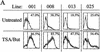
Restriction digestion analysis to determine the extent of DNA methylation over the transgene promoter. (A) Under a map of the reporter construct are illustrated the incidence of individual CpG dinucleotides and CCGG _Msp_I–_Hpa_II sites occuring in the sequence of the βA promoter, IL2R cDNA, and β/ε enhancer, showing the relatively CG-rich character of the promoter region. Indirect end labeling by digestion with _Xba_I and probing with IL2R cDNA detects a 867-bp fragment indicative of cutting at the site marked a. Cutting at any of a closely spaced quartet of sites at b produces indistinguishable indirect end-labeled bands of 961–1032 bp, and c gives a 1145-bp fragment. Very faint bands of higher molecular weight were only rarely seen. Digests were carried out overnight on 5 μg of DNA sample with 100 units of _Msp_I or _Hpa_II. Digestion by _Msp_I always went to completion. The extent of _Hpa_II digestion at the a site was quantitated for each _Hpa_II lane as the percentage of the intensity of the a band relative to the sum of the a, b, and c bands in the lane by PhosphoImager analysis. (B) DNA from sorted cells carrying uninsulated IL2R transgenes was digested with _Xba_I and _Msp_I (M) or _Hpa_II (H). These cells had been passaged 40–50 days in culture and dispalyed biphasic populations of IL2R activity in FACS analyses (see Fig. 2). (C) The same digests were performed on insulated IL2R lines following continuous passage in culture for >80 days, at which point all were still completely positive for IL2R activity.
Figure 6
Restriction digestion analysis to determine the extent of DNA methylation over the transgene promoter. (A) Under a map of the reporter construct are illustrated the incidence of individual CpG dinucleotides and CCGG _Msp_I–_Hpa_II sites occuring in the sequence of the βA promoter, IL2R cDNA, and β/ε enhancer, showing the relatively CG-rich character of the promoter region. Indirect end labeling by digestion with _Xba_I and probing with IL2R cDNA detects a 867-bp fragment indicative of cutting at the site marked a. Cutting at any of a closely spaced quartet of sites at b produces indistinguishable indirect end-labeled bands of 961–1032 bp, and c gives a 1145-bp fragment. Very faint bands of higher molecular weight were only rarely seen. Digests were carried out overnight on 5 μg of DNA sample with 100 units of _Msp_I or _Hpa_II. Digestion by _Msp_I always went to completion. The extent of _Hpa_II digestion at the a site was quantitated for each _Hpa_II lane as the percentage of the intensity of the a band relative to the sum of the a, b, and c bands in the lane by PhosphoImager analysis. (B) DNA from sorted cells carrying uninsulated IL2R transgenes was digested with _Xba_I and _Msp_I (M) or _Hpa_II (H). These cells had been passaged 40–50 days in culture and dispalyed biphasic populations of IL2R activity in FACS analyses (see Fig. 2). (C) The same digests were performed on insulated IL2R lines following continuous passage in culture for >80 days, at which point all were still completely positive for IL2R activity.
Figure 6
Restriction digestion analysis to determine the extent of DNA methylation over the transgene promoter. (A) Under a map of the reporter construct are illustrated the incidence of individual CpG dinucleotides and CCGG _Msp_I–_Hpa_II sites occuring in the sequence of the βA promoter, IL2R cDNA, and β/ε enhancer, showing the relatively CG-rich character of the promoter region. Indirect end labeling by digestion with _Xba_I and probing with IL2R cDNA detects a 867-bp fragment indicative of cutting at the site marked a. Cutting at any of a closely spaced quartet of sites at b produces indistinguishable indirect end-labeled bands of 961–1032 bp, and c gives a 1145-bp fragment. Very faint bands of higher molecular weight were only rarely seen. Digests were carried out overnight on 5 μg of DNA sample with 100 units of _Msp_I or _Hpa_II. Digestion by _Msp_I always went to completion. The extent of _Hpa_II digestion at the a site was quantitated for each _Hpa_II lane as the percentage of the intensity of the a band relative to the sum of the a, b, and c bands in the lane by PhosphoImager analysis. (B) DNA from sorted cells carrying uninsulated IL2R transgenes was digested with _Xba_I and _Msp_I (M) or _Hpa_II (H). These cells had been passaged 40–50 days in culture and dispalyed biphasic populations of IL2R activity in FACS analyses (see Fig. 2). (C) The same digests were performed on insulated IL2R lines following continuous passage in culture for >80 days, at which point all were still completely positive for IL2R activity.
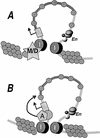
Figure 7
Two possible models of insulator action in protecting the reporter gene against formation of heterochromatin. In the first model (A), the insulator elements exclude methyl-CpG-binding proteins, histone deacetylases, or any other component required for formation of the silenced domain. Alternatively (B), the insulator elements are modeled as boundaries within which histone acetylase recruitment is preserved directly, thus favoring an acetylated, active domain over invading condensed chromatin. (I) Insulators; (P) promoter; (en) enhancer; (M/D) meCpG-binding protein/histone deacetylase complex; (A) histone acetylase.
Similar articles
- The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation.
Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. Mutskov VJ, et al. Genes Dev. 2002 Jun 15;16(12):1540-54. doi: 10.1101/gad.988502. Genes Dev. 2002. PMID: 12080092 Free PMC article. - Methylation of alpha-type embryonic globin gene alpha pi represses transcription in primary erythroid cells.
Singal R, vanWert JM, Ferdinand L Jr. Singal R, et al. Blood. 2002 Dec 1;100(12):4217-22. doi: 10.1182/blood-2002-02-0457. Epub 2002 Aug 1. Blood. 2002. PMID: 12393573 - Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation.
Schübeler D, Lorincz MC, Cimbora DM, Telling A, Feng YQ, Bouhassira EE, Groudine M. Schübeler D, et al. Mol Cell Biol. 2000 Dec;20(24):9103-12. doi: 10.1128/MCB.20.24.9103-9112.2000. Mol Cell Biol. 2000. PMID: 11094062 Free PMC article. - The insulation of genes from external enhancers and silencing chromatin.
Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G. Burgess-Beusse B, et al. Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16433-7. doi: 10.1073/pnas.162342499. Epub 2002 Aug 1. Proc Natl Acad Sci U S A. 2002. PMID: 12154228 Free PMC article. Review. - Chromatin structure and gene expression.
Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Felsenfeld G, et al. Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9384-8. doi: 10.1073/pnas.93.18.9384. Proc Natl Acad Sci U S A. 1996. PMID: 8790338 Free PMC article. Review.
Cited by
- Mi2β-mediated silencing of the fetal γ-globin gene in adult erythroid cells.
Amaya M, Desai M, Gnanapragasam MN, Wang SZ, Zu Zhu S, Williams DC Jr, Ginder GD. Amaya M, et al. Blood. 2013 Apr 25;121(17):3493-501. doi: 10.1182/blood-2012-11-466227. Epub 2013 Feb 26. Blood. 2013. PMID: 23444401 Free PMC article. - Efficient ROSA26-based conditional and/or inducible transgenesis using RMCE-compatible F1 hybrid mouse embryonic stem cells.
Haenebalcke L, Goossens S, Naessens M, Kruse N, Farhang Ghahremani M, Bartunkova S, Haigh K, Pieters T, Dierickx P, Drogat B, Nyabi O, Wirth D, Haigh JJ. Haenebalcke L, et al. Stem Cell Rev Rep. 2013 Dec;9(6):774-85. doi: 10.1007/s12015-013-9458-z. Stem Cell Rev Rep. 2013. PMID: 23877658 - Structural dynamics of nucleosomes at single-molecule resolution.
Choy JS, Lee TH. Choy JS, et al. Trends Biochem Sci. 2012 Oct;37(10):425-35. doi: 10.1016/j.tibs.2012.06.006. Epub 2012 Jul 23. Trends Biochem Sci. 2012. PMID: 22831768 Free PMC article. Review. - Stable expression and cell-specific chromatin structure of human alpha1-antitrypsin cosmid transgenes in rat hepatoma cells.
Rollini P, Xu L, Fournier RE. Rollini P, et al. Nucleic Acids Res. 2000 Sep 15;28(18):3605-14. doi: 10.1093/nar/28.18.3605. Nucleic Acids Res. 2000. PMID: 10982883 Free PMC article. - Lack of shielding of primer binding site silencer-mediated repression of an internal promoter in a retrovirus vector by the putative insulators scs, BEAD-1, and HS4.
Modin C, Pedersen FS, Duch M. Modin C, et al. J Virol. 2000 Dec;74(24):11697-707. doi: 10.1128/jvi.74.24.11697-11707.2000. J Virol. 2000. PMID: 11090169 Free PMC article.
References
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. - PubMed
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–604. - PubMed
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources