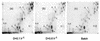Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool ("metabolome") analysis - PubMed (original) (raw)
Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool ("metabolome") analysis
H Tweeddale et al. J Bacteriol. 1998 Oct.
Abstract
Escherichia coli growing on glucose in minimal medium controls its metabolite pools in response to environmental conditions. The extent of pool changes was followed through two-dimensional thin-layer chromatography of all 14C-glucose labelled compounds extracted from bacteria. The patterns of metabolites and spot intensities detected by phosphorimaging were found to reproducibly differ depending on culture conditions. Clear trends were apparent in the pool sizes of several of the 70 most abundant metabolites extracted from bacteria growing in glucose-limited chemostats at different growth rates. The pools of glutamate, aspartate, trehalose, and adenosine as well as UDP-sugars and putrescine changed markedly. The data on pools observed by two-dimensional thin-layer chromatography were confirmed for amino acids by independent analysis. Other unidentified metabolites also displayed different spot intensities under various conditions, with four trend patterns depending on growth rate. As RpoS controls a number of metabolic genes in response to nutrient limitation, an rpoS mutant was also analyzed for metabolite pools. The mutant had altered metabolite profiles, but only some of the changes at slow growth rates were ascribable to the known control of metabolic genes by RpoS. These results indicate that total metabolite pool ("metabolome") analysis offers a means of revealing novel aspects of cellular metabolism and global regulation.
Figures
FIG. 1
The metabolome of E. coli growing on glucose. Extracts were obtained from chemostat-grown bacteria at low (a) or high (b) dilution rate with limiting 0.02% glucose or from an exponentially growing culture with 0.2% glucose (c). The extracts were subjected to TLC in solvent system A, and the phosphorimages were labelled with the positions of spots corresponding to the following: 1, glutamate; 2, trehalose; 3, glucose; 4, UDP-glucose plus UDP-galactose; 5, adenosine; 6, aspartate; 7, lysine; 8, UDP-_N_-acetylglucosamine; 9, glutathione; 10, putrescine. The lettered spots were unidentified compounds mentioned in the text. The origin of the chromatographs was the bottom left.
FIG. 2
The metabolome of E. coli growing on glucose and the effect of an rpoS mutation. The details are as for Fig. 1 except that the extracts were subject to TLC in solvent system B. Panels d and e represent the metabolomes of the rpoS mutant BW2996 growing at low (0.1 h−1) and high (0.6 h−1) dilution rates, respectively.
FIG. 3
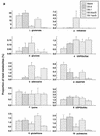
Changes in pool sizes as determined by metabolome analysis. The spots corresponding to the compounds identified (a) and unidentified (b) in Fig. 1 and 2 were quantitated with ImageQuant software in four to six independent determinations.
FIG. 3
Changes in pool sizes as determined by metabolome analysis. The spots corresponding to the compounds identified (a) and unidentified (b) in Fig. 1 and 2 were quantitated with ImageQuant software in four to six independent determinations.
FIG. 4
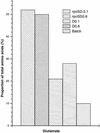
Glutamate pools measured by HPLC analysis. Duplicate samples of the five types of cell extract obtained as for Fig. 2 were analyzed for amino acids by using an AminoMate system. The mean of the glutamate quantities obtained for each type of sample is shown.
Similar articles
- New insights into the role of sigma factor RpoS as revealed in escherichia coli strains lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system.
Flores N, Escalante A, de Anda R, Báez-Viveros JL, Merino E, Franco B, Georgellis D, Gosset G, Bolívar F. Flores N, et al. J Mol Microbiol Biotechnol. 2008;14(4):176-92. doi: 10.1159/000109945. Epub 2007 Oct 15. J Mol Microbiol Biotechnol. 2008. PMID: 17938565 - Control of RpoS in global gene expression of Escherichia coli in minimal media.
Dong T, Schellhorn HE. Dong T, et al. Mol Genet Genomics. 2009 Jan;281(1):19-33. doi: 10.1007/s00438-008-0389-3. Epub 2008 Oct 9. Mol Genet Genomics. 2009. PMID: 18843507 - Hungry bacteria--definition and properties of a nutritional state.
Ferenci T. Ferenci T. Environ Microbiol. 2001 Oct;3(10):605-11. doi: 10.1046/j.1462-2920.2001.00238.x. Environ Microbiol. 2001. PMID: 11722540 Review. - The RpoS-mediated general stress response in Escherichia coli.
Battesti A, Majdalani N, Gottesman S. Battesti A, et al. Annu Rev Microbiol. 2011;65:189-213. doi: 10.1146/annurev-micro-090110-102946. Annu Rev Microbiol. 2011. PMID: 21639793 Free PMC article. Review.
Cited by
- Sequencing-guided re-estimation and promotion of cultivability for environmental bacteria.
Zheng M, Wen L, He C, Chen X, Si L, Li H, Liang Y, Zheng W, Guo F. Zheng M, et al. Nat Commun. 2024 Oct 20;15(1):9051. doi: 10.1038/s41467-024-53446-4. Nat Commun. 2024. PMID: 39426960 Free PMC article. - Combined proteomics and metabolomics analysis reveal the effect of a training course on the immune function of Chinese elite short-track speed skaters.
Li T, Shao J, An N, Chang Y, Xia Y, Han Q, Zhu F. Li T, et al. Immun Inflamm Dis. 2024 Oct;12(10):e70030. doi: 10.1002/iid3.70030. Immun Inflamm Dis. 2024. PMID: 39352112 Free PMC article. - RpoS and the bacterial general stress response.
Bouillet S, Bauer TS, Gottesman S. Bouillet S, et al. Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0015122. doi: 10.1128/mmbr.00151-22. Epub 2024 Feb 27. Microbiol Mol Biol Rev. 2024. PMID: 38411096 Review. - Plant Life with and without Oxygen: A Metabolomics Approach.
Yemelyanov VV, Puzanskiy RK, Shishova MF. Yemelyanov VV, et al. Int J Mol Sci. 2023 Nov 12;24(22):16222. doi: 10.3390/ijms242216222. Int J Mol Sci. 2023. PMID: 38003412 Free PMC article. Review. - Statistical methods and resources for biomarker discovery using metabolomics.
Anwardeen NR, Diboun I, Mokrab Y, Althani AA, Elrayess MA. Anwardeen NR, et al. BMC Bioinformatics. 2023 Jun 15;24(1):250. doi: 10.1186/s12859-023-05383-0. BMC Bioinformatics. 2023. PMID: 37322419 Free PMC article. Review.
References
- Apontoweil P, Berends W. Glutathione biosynthesis in Escherichia coli K 12. Properties of the enzymes and regulation. Biochim Biophys Acta. 1975;399:1–9. - PubMed
- Bailey J E. Toward a science of metabolic engineering. Science. 1991;252:1668–1674. - PubMed
- Bhattacharya M, Fuhrman L, Ingram A, Nickerson K W, Conway T. Single-run separation and detection of multiple metabolic intermediates by anion-exchange high-performance liquid chromatography and application to cell pool extracts prepared from Escherichia coli. Anal Biochem. 1995;232:98–106. - PubMed
- Bochner B R, Ames B N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials