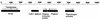Structural organization of virulence-associated plasmids of Yersinia pestis - PubMed (original) (raw)
Comparative Study
Structural organization of virulence-associated plasmids of Yersinia pestis
P Hu et al. J Bacteriol. 1998 Oct.
Abstract
The complete nucleotide sequence and gene organization of the three virulence plasmids from Yersinia pestis KIM5 were determined. Plasmid pPCP1 (9,610 bp) has a GC content of 45.3% and encodes two previously known virulence factors, an associated protein, and a single copy of IS100. Plasmid pCD1 (70,504 bp) has a GC content of 44.8%. It is known to encode a number of essential virulence determinants, regulatory functions, and a multiprotein secretory system comprising the low-calcium response stimulation that is shared with the other two Yersinia species pathogenic for humans (Y. pseudotuberculosis and Y. enterocolitica). A new pseudogene, which occurs as an intact gene in the Y. enterocolitica and Y. pseudotuberculosis-derived analogues, was found in pCD1. It corresponds to that encoding the lipoprotein YlpA. Several intact and partial insertion sequences and/or transposons were also found in pCD1, as well as six putative structural genes with high homology to proteins of unknown function in other yersiniae. The sequences of the genes involved in the replication of pCD1 are highly homologous to those of the cognate plasmids in Y. pseudotuberculosis and Y. enterocolitica, but their localization within the plasmid differs markedly from those of the latter. Plasmid pMT1 (100,984 bp) has a GC content of 50.2%. It possesses two copies of IS100, which are located 25 kb apart and in opposite orientations. Adjacent to one of these IS100 inserts is a partial copy of IS285. A single copy of an IS200-like element (recently named IS1541) was also located in pMT1. In addition to 5 previously described genes, such as murine toxin, capsule antigen, capsule anchoring protein, etc., 30 homologues to genes of several bacterial species were found in this plasmid, and another 44 open reading frames without homology to any known or hypothetical protein in the databases were predicted.
Figures
FIG. 1
Structural organization of the 9,610-bp plasmid pPCP1 derived from Y. pestis KIM5. BLAST searches using the entire nucleotide sequence obtained in this work were performed to precisely localize potential new ORFs, insertion sequence elements, and the three previously described genes present in pPCP. The directions of transcription of these genes are indicated by the arrows. The single IS_100_ element was used to define position 1 of this plasmid. The characteristics of the genes and proteins involved are described in the text. The numbering above the line is the molecular size in base pairs.
FIG. 2
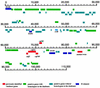
Physical map and genetic organization of pCD1. ORFs, insertion sequences, and other genetic elements were located in the map by using BLAST searches and GeneMark. ORFs and genes in the map are color coded according to function or unique characteristic, and their designations are placed either above or below the colored bars. The scale indicates the number of nucleotides measured from the start of the single IS_100_ found in this plasmid. Genes in the figures are located precisely in the map and drawn to scale directly from sequence annotation by using an in-house, UNIX-based annotation-rendering program. Genes positioned on top of each line are transcribed from left to right, whereas those placed below the line are encoded by the complementary strand. The two pseudogenes (ylpA and yadA) are represented by partially colored bars. The position of the origin of replication is marked as oriR. The characteristics of the genes, proteins, and sequences depicted are described in the text and in Table 2.
FIG. 3
Physical map and genetic organization of pMT1. ORFs, genes, and other features displayed in the map are depicted as described in Fig. 2. The characteristics of all of the elements described in the map are defined in the text and in Table 3. The caf1A and caf1 genes located at about 70,000 nucleotides are incorrectly labeled calf1A and calf1, respectively.
FIG. 4
Diagram comparing the organization of selected genes and elements of the Lcr plasmid in yersiniae. Shown are circular maps of the pCD1 plasmid and of the homologous pYV and pCad plasmids derived from Y. enterocolitica and Y. pseudotuberculosis, respectively. The relative positions of selected loci with respect to the origin of replication of pCD1 are shown. Outer circle, pCD1; middle circle, pIB1 (Y. pseudotuberculosis); inner circle, pYVe O:9 (Y. enterocolitica). The nomenclature and approximate positions of genes in pYV and pIB1 are from Iriarte and Cornelis (22), Persson et al. (38), and Salyers and Whitt (51). The genes and sequence features of pCD1 and the corresponding regions in pYV and pIB1 are depicted in the same color to aid in their visualization (e.g., the repBA, oriR, and ypkA regions are presented in green, black, and red, respectively). Numbering inside the circles indicates the approximate sizes of the plasmids in nucleotides, measured from the start of their origins of replication. Arrows above each color segment representing a gene or gene group point to the direction of transcription.
Similar articles
- DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5.
Perry RD, Straley SC, Fetherston JD, Rose DJ, Gregor J, Blattner FR. Perry RD, et al. Infect Immun. 1998 Oct;66(10):4611-23. doi: 10.1128/IAI.66.10.4611-4623.1998. Infect Immun. 1998. PMID: 9746557 Free PMC article. - Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen.
Lindler LE, Plano GV, Burland V, Mayhew GF, Blattner FR. Lindler LE, et al. Infect Immun. 1998 Dec;66(12):5731-42. doi: 10.1128/IAI.66.12.5731-5742.1998. Infect Immun. 1998. PMID: 9826348 Free PMC article. - Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100.
Podladchikova ON, Dikhanov GG, Rakin AV, Heesemann J. Podladchikova ON, et al. FEMS Microbiol Lett. 1994 Sep 1;121(3):269-74. doi: 10.1111/j.1574-6968.1994.tb07111.x. FEMS Microbiol Lett. 1994. PMID: 7926680 - The yersiniae--a model genus to study the rapid evolution of bacterial pathogens.
Wren BW. Wren BW. Nat Rev Microbiol. 2003 Oct;1(1):55-64. doi: 10.1038/nrmicro730. Nat Rev Microbiol. 2003. PMID: 15040180 Review. - The plasmid-encoded outer-membrane proteins of Yersinia pestis.
Straley SC. Straley SC. Rev Infect Dis. 1988 Jul-Aug;10 Suppl 2:S323-6. doi: 10.1093/cid/10.supplement_2.s323. Rev Infect Dis. 1988. PMID: 3055200 Review.
Cited by
- Exploring lateral genetic transfer among microbial genomes using TF-IDF.
Cong Y, Chan YB, Ragan MA. Cong Y, et al. Sci Rep. 2016 Jul 25;6:29319. doi: 10.1038/srep29319. Sci Rep. 2016. PMID: 27452976 Free PMC article. - Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases.
Keim PS, Wagner DM. Keim PS, et al. Nat Rev Microbiol. 2009 Nov;7(11):813-21. doi: 10.1038/nrmicro2219. Epub 2009 Oct 12. Nat Rev Microbiol. 2009. PMID: 19820723 Free PMC article. Review. - Reverse line blot macroarray for simultaneous detection and characterization of four biological warfare agents.
Vanlalhmuaka, Thavachelvam K, Tuteja U, Sarika K, Nagendra S, Kumar S. Vanlalhmuaka, et al. Indian J Microbiol. 2013 Mar;53(1):41-7. doi: 10.1007/s12088-012-0330-7. Epub 2012 Nov 6. Indian J Microbiol. 2013. PMID: 24426077 Free PMC article. - Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies.
Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R. Healy FG, et al. J Bacteriol. 2002 Apr;184(7):2019-29. doi: 10.1128/JB.184.7.2019-2029.2002. J Bacteriol. 2002. PMID: 11889110 Free PMC article. - The virulence plasmid of Yersinia, an antihost genome.
Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. Cornelis GR, et al. Microbiol Mol Biol Rev. 1998 Dec;62(4):1315-52. doi: 10.1128/MMBR.62.4.1315-1352.1998. Microbiol Mol Biol Rev. 1998. PMID: 9841674 Free PMC article. Review.
References
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Ben-Gurion R, Shafferman A. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid. 1981;5:183–187. - PubMed
- Bodenteich A, Chissoe S, Wang Y-F, Roe B A. Shotgun cloning as the strategy of choice to generate templates for high-throughput dideoxynucleotide sequencing. In: Adams M, Fields C, Venter J C, editors. Automated DNA sequencing and analysis. London, England: Academic Press; 1994. pp. 42–49.
- Borodovsky M, Koonin E V, Rudd K E. New genes in old sequences: a strategy for finding genes in the bacterial genome. Trends Biochem Sci. 1994;19:309–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous