Bias in template-to-product ratios in multitemplate PCR - PubMed (original) (raw)
Comparative Study
Bias in template-to-product ratios in multitemplate PCR
M F Polz et al. Appl Environ Microbiol. 1998 Oct.
Abstract
Bias introduced by the simultaneous amplification of specific genes from complex mixtures of templates remains poorly understood. To explore potential causes and the extent of bias in PCR amplification of 16S ribosomal DNAs (rDNAs), genomic DNAs of two closely and one distantly related bacterial species were mixed and amplified with universal, degenerate primers. Quantification and comparison of template and product ratios showed that there was considerable and reproducible overamplification of specific templates. Variability between replicates also contributed to the observed bias but in a comparatively minor way. Based on these initial observations, template dosage and differences in binding energies of permutations of the degenerate, universal primers were tested as two likely causes of this template-specific bias by using 16S rDNA templates modified by site-directed mutagenesis. When mixtures of mutagenized templates containing AT- and GC-rich priming sites were used, templates containing the GC-rich permutation amplified with higher efficiency, indicating that different primer binding energies may to a large extent be responsible for overamplification. In contrast, gene copy number was found to be an unlikely cause of the observed bias. Similarly, amplification from DNA extracted from a natural community to which different amounts of genomic DNA of a single bacterial species were added did not affect relative product ratios. Bias was reduced considerably by using high template concentrations, by performing fewer cycles, and by mixing replicate reaction preparations.
Figures
FIG. 1
Dot blot analyses showing the specificity of the oligonucleotide probes for their targets. (A) Genomic DNAs (left dots) and PCR-amplified 16S rDNAs (right dots) of B. subtilis, V. fischeri, and V. anguillarum were blotted together on three replicate membranes and hybridized with the specific probes Bsu, Van, and Vfi, respectively. (B) PCR-amplified 16S rDNAs of mutagenized plasmids Eco(GC), Eco(AT), and Eco(AT)m were blotted on two separate membranes and hybridized with the specific probes Eco and EcoM, respectively. The electronic image was taken from X-ray film exposed for 5 h.
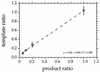
FIG. 2
Effect of gene dosage as determined by amplification with different template ratios of Eco(AT) and Eco(AT)m and quantification of product ratios by quantitative dot blotting with probes Eco and EcoM. Regression analysis showed that the relationship between template and product ratios was linear. The vertical bars indicate standard deviations.
FIG. 3
Alignment of the sequences of universal amplification primers 27F and 1492R and their target regions on the 16S rRNA genes of B. subtilis, V. fischeri, and V. anguillarum. The two primers each contain a single degeneracy (between C and T and between C and A, respectively). In the B. subtilis gene both priming sites contain a G at the degenerate site, which most likely results in a higher melting temperature for the primer-target duplex than the melting temperature for the two Vibrio genes, which contain an A and a T at the two positions.
Similar articles
- Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR.
Suzuki MT, Giovannoni SJ. Suzuki MT, et al. Appl Environ Microbiol. 1996 Feb;62(2):625-30. doi: 10.1128/aem.62.2.625-630.1996. Appl Environ Microbiol. 1996. PMID: 8593063 Free PMC article. - PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains.
Yamamoto S, Harayama S. Yamamoto S, et al. Appl Environ Microbiol. 1995 Mar;61(3):1104-9. doi: 10.1128/aem.61.3.1104-1109.1995. Appl Environ Microbiol. 1995. PMID: 7793912 Free PMC article. - Metagenome complexity and template length are the main causes of bias in PCR-based bacteria community analysis.
Peng W, Li X, Wang C, Cao H, Cui Z. Peng W, et al. J Basic Microbiol. 2018 Nov;58(11):987-997. doi: 10.1002/jobm.201800265. Epub 2018 Aug 9. J Basic Microbiol. 2018. PMID: 30091475 - [Polymerase chain reaction, cold probes and clinical diagnosis].
Haras D, Amoros JP. Haras D, et al. Sante. 1994 Jan-Feb;4(1):43-52. Sante. 1994. PMID: 7909267 Review. French. - Saturation mutagenesis by mutagenic oligonucleotide-directed PCR amplification (Mod-PCR).
Chiang LW. Chiang LW. Methods Mol Biol. 1996;57:311-21. doi: 10.1385/0-89603-332-5:311. Methods Mol Biol. 1996. PMID: 8850017 Review. No abstract available.
Cited by
- Genotypic and phenotypic characterization of the first Latin America isolates of Corynebacterium rouxii, a recently described member of the Corynebacterium diphtheriae complex reported in Europe.
de Oliveira Sant'Anna L, Dos Santos LS, Ramos JN, Bokermann S, Bernardes Sousa MÂ, Prates FD, Mattos-Guaraldi AL, Vieira VV, Araújo MRB. de Oliveira Sant'Anna L, et al. Braz J Microbiol. 2024 Oct 7. doi: 10.1007/s42770-024-01526-4. Online ahead of print. Braz J Microbiol. 2024. PMID: 39373943 - The unique chemical and microbiological signatures of an array of bottled drinking water.
Nadreen YM, Vrouwenvelder JS, Saikaly PE, Gonzalez-Gil G. Nadreen YM, et al. Front Microbiol. 2024 Sep 16;15:1441142. doi: 10.3389/fmicb.2024.1441142. eCollection 2024. Front Microbiol. 2024. PMID: 39351306 Free PMC article. - Quantitating primer-template interactions using deconstructed PCR.
Kahsen J, Sherwani SK, Naqib A, Jeon T, Wu LYA, Green SJ. Kahsen J, et al. PeerJ. 2024 Aug 8;12:e17787. doi: 10.7717/peerj.17787. eCollection 2024. PeerJ. 2024. PMID: 39131619 Free PMC article. - Investigating the lignocellulolytic gut microbiome of huhu grubs (Prionoplus reticularis) using defined diets and dietary switch.
Viswam J, Baptista M, Lee CK, Morgan H, McDonald IR. Viswam J, et al. PeerJ. 2024 Jul 2;12:e17597. doi: 10.7717/peerj.17597. eCollection 2024. PeerJ. 2024. PMID: 38974417 Free PMC article.
References
- Arnheim N, Erlich H. Polymerase chain reaction strategy. Annu Rev Biochem. 1992;61:131–156. - PubMed
- Chandler D P, Fredrickson J K, Brockman F J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol. 1997;6:475–483. - PubMed
- Erlich H A, Gelfand D, Sninsky J J. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous