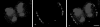Transcription sites are not correlated with chromosome territories in wheat nuclei - PubMed (original) (raw)
Transcription sites are not correlated with chromosome territories in wheat nuclei
R Abranches et al. J Cell Biol. 1998.
Abstract
We have determined the relationship between overall nuclear architecture, chromosome territories, and transcription sites within the nucleus, using three-dimensional confocal microscopy of well preserved tissue sections of wheat roots. Chromosome territories were visualized by GISH using rye genomic probe in wheat/rye translocation and addition lines. The chromosomes appeared as elongated regions and showed a clear centromere-telomere polarization, with the two visualized chromosomes lying approximately parallel to one another across the nucleus. Labeling with probes to telomeres and centromeres confirmed a striking Rabl configuration in all cells, with a clear clustering of the centromeres, and cell files often maintained a common polarity through several division cycles. Transcription sites were detected by BrUTP incorporation in unfixed tissue sections and revealed a pattern of numerous foci uniformly distributed throughout the nucleoplasm, as well as more intensely labeled foci in the nucleoli. It has been suggested that the gene-rich regions in wheat chromosomes are clustered towards the telomeres. However, we found no indication of a difference in concentration of transcription sites between telomere and centromere poles of the nucleus. Neither could we detect any evidence that the transcription sites were preferentially localized with respect to the chromosome territorial boundaries.
Figures
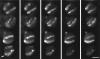
Figure 1
Root tissue from wheat 1R addition line, in which a pair of rye chromosomes (1R) is present. The rye chromosomes have been labeled by genomic fluorescence in situ hybridization using a total rye genomic DNA probe. A series of optical sections collected by confocal microscopy is shown. The chromosomes stretch across the nuclei, the two arms next to each other, and the two labeled chromosomes are usually parallel to one another. In some cases, the two arms can be distinguished (arrow). Focal distance between section = 1 μm. Bar, 10 μm.
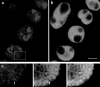
Figure 2
(a) Wheat root tissue double labeled by fluorescence in situ hybridization with probes to the centromeres (green) and telomeres (red). Projection of five confocal optical sections (focal distance between original sections = 1 μm). (b) Diagram showing the interpretation of the labeling in (a) as the Rabl configuration. The chromosomes must all be parallel to one another, with the centromeres clustered on one side of the nuclear periphery, and the telomeres somewhat more dispersed on the other side of the nuclear periphery. A common (alternating) polarity is often maintained through the lines of cells as in this image. Bar, 10 μm.
Figure 3
Labeling of transcription sites in wheat root tissue by incorporation of BrUTP. A single confocal optical section is shown. (a) BrUTP labeling of several nuclei (red). The nucleolar transcription sites are more strongly labeled than the nucleoplasmic sites, and are overexposed in this image. The nucleoplasmic sites are fairly uniformly distributed throughout the nucleoplasm. There is some variability in the intensity of BrUTP labeling, so that, for example, the upper left nucleus is less strongly labeled, but the pattern of labeling is equivalent to that in the other cells. (b) Corresponding DAPI image (blue). (c) Enlargement of the boxed area in a, showing the correspondence between BrUTP sites and chromatin structure. In some cases transcription foci localize to DAPI-dark regions (e.g., arrows), but this is not universal. Bar, 10 μm.
Figure 4
A single confocal image showing double labeling of transcription sites (red, left panel) and centromeres (green, central panel). The two labels are superimposed in the right panel. There is no indication of a polarized distribution of transcription sites, which appear as dense at the centromeric poles of the nuclei as at the opposite, telomeric poles. Bar, 10 μm.
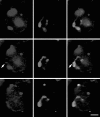
Figure 5
Double labeling of the wheat 1A/1R translocation line, in which one arm of wheat chromosome 1A is substituted by an arm of rye chromosome 1. BrUTP incorporation is shown in red (left-hand panels), genomic in situ labeling in green (central panels), the two labels superimposed in the right hand panels. Three consecutive confocal sections are shown. The distribution of transcription sites shows no sign of being excluded from the interior of the labeled chromosome territory. In fact two prominent sites are clearly inside the chromosome territory (arrows). Section spacing = 1 μm. Bar, 5 μm.
Similar articles
- Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells.
Dong F, Jiang J. Dong F, et al. Chromosome Res. 1998 Nov;6(7):551-8. doi: 10.1023/a:1009280425125. Chromosome Res. 1998. PMID: 9886774 - Chromosome arrangement and behaviour of two rye homologous telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus.
Maestra B, Hans de Jong J, Shepherd K, Naranjo T. Maestra B, et al. Chromosome Res. 2002;10(8):655-67. doi: 10.1023/a:1021564327226. Chromosome Res. 2002. PMID: 12575794 - The nucleus: a highly organized but dynamic structure.
Gonzalez-Melendi P, Beven A, Boudonck K, Abranches R, Wells B, Dolan L, Shaw P. Gonzalez-Melendi P, et al. J Microsc. 2000 Jun;198(Pt 3):199-207. doi: 10.1046/j.1365-2818.2000.00701.x. J Microsc. 2000. PMID: 10849198 - The architecture of interphase chromosomes and nucleolar transcription sites in plants.
Shaw PJ, Abranches R, Paula Santos A, Beven AF, Stoger E, Wegel E, González-Melendi P. Shaw PJ, et al. J Struct Biol. 2002 Oct-Dec;140(1-3):31-8. doi: 10.1016/s1047-8477(02)00537-3. J Struct Biol. 2002. PMID: 12490151 Review. - Organization and dynamics of plant interphase chromosomes.
Schubert I, Shaw P. Schubert I, et al. Trends Plant Sci. 2011 May;16(5):273-81. doi: 10.1016/j.tplants.2011.02.002. Epub 2011 Mar 9. Trends Plant Sci. 2011. PMID: 21393049 Review.
Cited by
- Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes.
Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I. Weierich C, et al. Chromosome Res. 2003;11(5):485-502. doi: 10.1023/a:1025016828544. Chromosome Res. 2003. PMID: 12971724 - Wheat chromatin architecture is organized in genome territories and transcription factories.
Concia L, Veluchamy A, Ramirez-Prado JS, Martin-Ramirez A, Huang Y, Perez M, Domenichini S, Rodriguez Granados NY, Kim S, Blein T, Duncan S, Pichot C, Manza-Mianza D, Juery C, Paux E, Moore G, Hirt H, Bergounioux C, Crespi M, Mahfouz MM, Bendahmane A, Liu C, Hall A, Raynaud C, Latrasse D, Benhamed M. Concia L, et al. Genome Biol. 2020 Apr 29;21(1):104. doi: 10.1186/s13059-020-01998-1. Genome Biol. 2020. PMID: 32349780 Free PMC article. - Plant 3D Chromatin Organization: Important Insights from Chromosome Conformation Capture Analyses of the Last 10 Years.
Zhang X, Wang T. Zhang X, et al. Plant Cell Physiol. 2021 Dec 10;62(11):1648-1661. doi: 10.1093/pcp/pcab134. Plant Cell Physiol. 2021. PMID: 34486654 Free PMC article. Review. - Spatial relationship between transcription sites and chromosome territories.
Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Verschure PJ, et al. J Cell Biol. 1999 Oct 4;147(1):13-24. doi: 10.1083/jcb.147.1.13. J Cell Biol. 1999. PMID: 10508851 Free PMC article.
References
- Aragón-Alcaide L, Reader S, Beven A, Shaw P, Miller T, Moore G. Association of homologous chromosomes during floral development. Curr Biol. 1997;7:905–908. - PubMed
- Avivi L, Feldman M. Arrangement of chromosomes in the interphase nucleus of plants. Hum Genet. 1980;55:281–295. - PubMed
- Beven AF, Lee R, Razaz M, Leader DJ, Brown JWS, Shaw PJ. The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J Cell Sci. 1996;109:1241–1251. - PubMed
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes ar centromeric heterochromatin. Cell. 1997;91:845–854. - PubMed