Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae - PubMed (original) (raw)
Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae
M Oakes et al. J Cell Biol. 1998.
Abstract
The nucleolus in Saccharomyces cerevisiae is a crescent-shaped structure that makes extensive contact with the nuclear envelope. In different chromosomal rDNA deletion mutants that we have analyzed, the nucleolus is not organized into a crescent structure, as determined by immunofluorescence microscopy, fluorescence in situ hybridization, and electron microscopy. A strain carrying a plasmid with a single rDNA repeat transcribed by RNA polymerase I (Pol I) contained a fragmented nucleolus distributed throughout the nucleus, primarily localized at the nuclear periphery. A strain carrying a plasmid with the 35S rRNA coding region fused to the GAL7 promoter and transcribed by Pol II contained a rounded nucleolus that often lacked extensive contact with the nuclear envelope. Ultrastructurally distinct domains were observed within the round nucleolus. A similar rounded nucleolar morphology was also observed in strains carrying the Pol I plasmid in combination with mutations that affect Pol I function. In a Pol I-defective mutant strain that carried copies of the GAL7-35S rDNA fusion gene integrated into the chromosomal rDNA locus, the nucleolus exhibited a round morphology, but was more closely associated with the nuclear envelope in the form of a bulge. Thus, both the organization of the rDNA genes and the type of polymerase involved in rDNA expression strongly influence the organization and localization of the nucleolus.
Figures
Figure 7
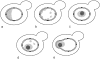
Schematic representation of nucleolar morphologies in yeast mutants. (a) Crescent-shaped nucleolus observed in wild-type cells. (b) Dispersed nucleolus observed in cells deleted for rDNA and bearing rDNA on a multicopy plasmid. (c) Condensed nucleolus observed in cells deleted for rDNA and bearing a multicopy plasmid expressing rDNA from a Pol II promoter. (d) Nucleolus observed in an rpa135Δ mutant that lacks functional Pol I and carries a multicopy plasmid that expresses rDNA from a Pol II promoter. (e) Nuclear envelope associated nucleolus observed in an rpa135Δ mutant containing 20–25 repeats at the rDNA locus that express rDNA from a Pol II promoter. The panels a–e correspond to EM images obtained with strains: NOY505 (a); NOY758 and NOY770 (b); NOY759, NOY773, and NOY777 (c); NOY408-1a (d); YJV100 (e). Characteristics of these strains are summarized in Table II.
Figure 4
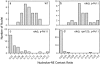
Morphometric analysis of the extent of contact between the nucleolus and the nuclear envelope. The nucleolus– nuclear envelope (NE) contact ratio is the linear distance of contact between the nucleolus and nuclear envelope divided by the cross-sectional area of the nucleolus (see Materials and Methods). A low value indicates less contact of the nucleolus with the nuclear envelope. The distribution of ratios is shown for NOY505 (control; panel a), NOY758 (rdnΔ, pRDN-hyg1; panel b), NOY759 (rdnΔ, pNOY353; panel c), and NOY777 (rdnΔ, rpa12Δ, pRDN-hyg1; panel d). The number of individual nuclei analyzed from each strain was (n =) 28, 27, 32, and 32, respectively. Panels a–d correspond to a–d in Fig. 3.
Figure 3
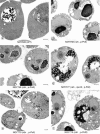
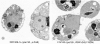
EM analysis of yeast mutants in which chromosomal rDNA repeats are deleted. Yeast strains NOY505 (control), NOY758 (rdnΔ, pRDN-hyg1), NOY759 (rdnΔ, pNOY353), NOY777 (rdnΔ, rpa12Δ, pRDN-hyg1), NOY770 (rdnΔΔ, pRDN-hyg1), and NOY773 (rdnΔΔ, pNOY353) were grown in YEP–galactose at 25°C to an A 600 of ∼0.5 and prepared for EM as described in Materials and Methods. Representative fields of cells in which nuclei and nucleoli are visible are shown. The electron-dense material representing the normal nucleolus is located at the lower portion of the two nuclei shown in a, NOY505, WT. The nuclear envelope is marked with arrows to serve as a point of reference. V, vacuole. Bars, 0.5 μm.
Figure 1
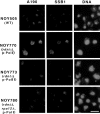
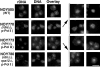
IFM analysis of nucleolar proteins in yeast mutants in which the chromosomal rDNA repeats are deleted. Yeast strains NOY505 (control), NOY770 (rdnΔΔ, pRDN-hyg1), NOY773 (rdnΔΔ, pNOY353) and NOY780 (rdnΔΔ, rpa12Δ, pRDN-hyg1) were analyzed for the Pol I A190 subunit, nucleolar protein Ssb1p, and DNA as described in Materials and Methods. Bar, 5 μm.
Figure 2
FISH analysis of rDNA in yeast mutants in which chromosomal rDNA repeats are deleted. Yeast strains NOY505 (control), NOY770 (rdnΔΔ, pRDN-hyg1), NOY773 (rdnΔΔ, pNOY353) and NOY780 (rdnΔΔ, rpa12Δ, pRDN-hyg1) were analyzed for rDNA and DNA as described in Materials and Methods. Additional images of NOY770 and NOY773 are indicated in brackets. Images of rDNA and DNA were pseudocolored red and blue, respectively. Individual images were overlayed and regions of colocalization appear as pink staining. Bar, 5 μm.
Figure 5
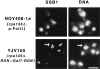
IFM analysis of nucleolar structures in strains transcribing the GAL7-35S rDNA fusion gene on a plasmid (NOY408-1a) or the same fusion gene integrated into chromosome XII at the RDN locus (YJV100) by use of Pol II. Nucleolar protein Ssb1p was analyzed by IFM and DNA was stained by DAPI as described in Materials and Methods. Arrows indicate an example of the nucleolus as revealed by anti-Ssb1p that shows a special structure protruding from the main body of the nucleus. Bar, 5 μm.
Figure 6
EM analysis of yeast mutants. Yeast strains NOY408-1a (rpa135Δ, p-Pol II) (a) and YJV100 (RDN::GAL7-RDN) (b) were analyzed as described in Fig. 3. Representative fields of cells in which nuclei and nucleoli are visible are shown. The nuclear envelope is marked with arrows as a point of reference. SPB, spindle pole body. Bars, 0.5 μm.
Similar articles
- Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo.
Wai HH, Vu L, Oakes M, Nomura M. Wai HH, et al. Nucleic Acids Res. 2000 Sep 15;28(18):3524-34. doi: 10.1093/nar/28.18.3524. Nucleic Acids Res. 2000. PMID: 10982872 Free PMC article. - Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I.
Oakes M, Nogi Y, Clark MW, Nomura M. Oakes M, et al. Mol Cell Biol. 1993 Apr;13(4):2441-55. doi: 10.1128/mcb.13.4.2441-2455.1993. Mol Cell Biol. 1993. PMID: 8455621 Free PMC article. - Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants.
Trumtel S, Léger-Silvestre I, Gleizes PE, Teulières F, Gas N. Trumtel S, et al. Mol Biol Cell. 2000 Jun;11(6):2175-89. doi: 10.1091/mbc.11.6.2175. Mol Biol Cell. 2000. PMID: 10848637 Free PMC article. - The nucleolus.
Schwarzacher HG, Wachtler F. Schwarzacher HG, et al. Anat Embryol (Berl). 1993 Dec;188(6):515-36. doi: 10.1007/BF00187008. Anat Embryol (Berl). 1993. PMID: 8129175 Review. - Multiparameter microscopic analysis of nucleolar structure and ribosomal gene transcription.
Trendelenburg MF, Zatsepina OV, Waschek T, Schlegel W, Tröster H, Rudolph D, Schmahl G, Spring H. Trendelenburg MF, et al. Histochem Cell Biol. 1996 Aug;106(2):167-92. doi: 10.1007/BF02484399. Histochem Cell Biol. 1996. PMID: 8877378 Review.
Cited by
- The concept of self-organization in cellular architecture.
Misteli T. Misteli T. J Cell Biol. 2001 Oct 15;155(2):181-5. doi: 10.1083/jcb.200108110. Epub 2001 Oct 15. J Cell Biol. 2001. PMID: 11604416 Free PMC article. Review. - Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast.
Oakes ML, Siddiqi I, French SL, Vu L, Sato M, Aris JP, Beyer AL, Nomura M. Oakes ML, et al. Mol Cell Biol. 2006 May;26(10):3889-901. doi: 10.1128/MCB.26.10.3889-3901.2006. Mol Cell Biol. 2006. PMID: 16648483 Free PMC article. - Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase.
Wang BD, Yong-Gonzalez V, Strunnikov AV. Wang BD, et al. Cell Cycle. 2004 Jul;3(7):960-7. doi: 10.4161/cc.3.7.1003. Epub 2004 Jul 4. Cell Cycle. 2004. PMID: 15190202 Free PMC article. - Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing.
Fath S, Milkereit P, Podtelejnikov AV, Bischler N, Schultz P, Bier M, Mann M, Tschochner H. Fath S, et al. J Cell Biol. 2000 May 1;149(3):575-90. doi: 10.1083/jcb.149.3.575. J Cell Biol. 2000. PMID: 10791972 Free PMC article.
References
- Bourgeois CA, Hubert J. Spatial relationship between the nucleolus and the nuclear envelope: structural aspects and functional significance. Int Rev Cytol. 1988;111:1–52. - PubMed
- Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. - PubMed
- Bryk M, Banjeree M, Murphy M, Knidsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1locus of yeast. Genes Dev. 1997;11:255–269. - PubMed
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. - PubMed
- Castano IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topisomerase I in Saccharomyces cerevisiae. . Genes Dev. 1996;10:2564–2576. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases