Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1 - PubMed (original) (raw)
Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1
G K Chan et al. J Cell Biol. 1998.
Abstract
We have identified a 350-amino acid domain in the kinetochore motor CENP-E that specifies kinetochore binding in mitosis but not during interphase. The kinetochore binding domain was used in a yeast two-hybrid screen to isolate interacting proteins that included the kinetochore proteins CENP-E, CENP-F, and hBUBR1, a BUB1-related kinase that was found to be mutated in some colorectal carcinomas (Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K. Wilson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Nature. 392:300-303). CENP-F, hBUBR1, and CENP-E assembled onto kinetochores in sequential order during late stages of the cell cycle. These proteins therefore define discrete steps along the kinetochore assembly pathway. Kinetochores of unaligned chromosome exhibited stronger hBUBR1 and CENP-E staining than those of aligned chromosomes. CENP-E and hBUBR1 remain colocalized at kinetochores until mid-anaphase when hBUBR1 localized to portions of the spindle midzone that did not overlap with CENP-E. As CENP-E and hBUBR1 can coimmunoprecipitate with each other from HeLa cells, they may function as a motor-kinase complex at kinetochores. However, the complex distribution pattern of hBUBR1 suggests that it may regulate multiple functions that include the kinetochore and the spindle midzone.
Figures
Figure 1
Expression of proA and gfp:CENP-E fusion proteins in HeLa cells. (A) Schematic diagram depicting the overlapping set of CENP-E cDNA fragments that were tested for their ability to bind kinetochores (+) or not (−). Hatched box, the kinetochore binding domain. (B) Immunoblot of transfected cell lysates expressing various proA:CENP-E fusion proteins as detected using alkaline phosphatase–conjugated human IgG. Gfp:CENP-E fusion proteins were detected with rat anti-gfp antibodies.
Figure 1
Expression of proA and gfp:CENP-E fusion proteins in HeLa cells. (A) Schematic diagram depicting the overlapping set of CENP-E cDNA fragments that were tested for their ability to bind kinetochores (+) or not (−). Hatched box, the kinetochore binding domain. (B) Immunoblot of transfected cell lysates expressing various proA:CENP-E fusion proteins as detected using alkaline phosphatase–conjugated human IgG. Gfp:CENP-E fusion proteins were detected with rat anti-gfp antibodies.
Figure 2
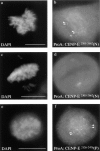
Localization of CENP-E kinetochore binding domain. (a) Gfp:CENP-E1558–2662(F) binds kinetochores and colocalizes with ACA but not with microtubules. (b) Gfp:CENP-E1958–2662(J) colocalizes with ACA but not with microtubules. (c) Gfp:CENP-E803–2123(E) is expressed but not localized to kinetochores. ACA staining was visualized with Cy-5 anti–human secondary antibodies, anti-tubulin antibodies were detected with biotinylated anti–mouse secondary and Texas red conjugated to streptavidin. Gfp fusions were visualized in the FITC channel. (d) Kinetochores containing gfp:CENP-E1958–2662(J) were probed for endogenous CENP-E using a rabbit anti–CENP-E “neck” antibody (Schaar et al., 1997) and compared with a neighboring untransfected metaphase that was stained with the same antibody. (e) Kinetochores containing gfp:CENP-E1958–2662(J) probed with CENP-F and compared with an untransfected metaphase cell also probed with CENP-F. Rabbit anti–CENP-E and anti–CENP-F antibodies were detected with Texas red–conjugated anti–rabbit secondary antibodies. Chromosomes were stained with DAPI. All samples were first fixed and then permeabilized. Bars, 10 μm.
Figure 3
Localization of the minimal kinetochore binding domain. proA:CENP-E2305–2662(N) binds to all kinetochores in one cell (b) but only a few in another (d). (f) The minimal kinetochore binding domain lies within proA:CENP-E2126–2476(P). ProA:CENP-E fusions were visualized with Texas red conjugated to human IgG (b, d, and f). Chromosomes are stained with DAPI (a, c, and e). Arrowheads, double-dot kinetochore staining. All samples were first fixed and then permeabilized. Bars, 10 μm.
Figure 4
Ectopic expression of CENP-E in interphase nuclei. (a) HeLa lysates express proA: CENP-E2126–2565:NLS(O). Immunoblot was probed with alkaline phosphatase–conjugated human IgG. (b–d) HeLa cells cotransfected with (c) proA:CENP-E2126–2565:NLS(O) and (d) CENP-B:gfp. (e and f) ProA:CENP-E2126–2565: NLS(O) localize to kinetochores during mitosis. (c and f) ProA:CENP-E2126–2565:NLS(O) was visualized with Texas red–conjugated human IgG. (b and e) Chromosomes and nuclei were visualized with DAPI. Arrowheads, double-dot kinetochore staining. All samples were first fixed and then permeabilized. Bars, 10 μm.
Figure 5
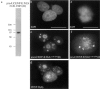
Identification of human hBUBR1 kinase. (a) Dendrogram depicting the relatedness of yeast, mouse, rat (partial clone), and human BUB1 kinases was generated with GCG PILEUP program. (b) Alignment of the NH2-terminal portion of mBUB1, hBUBR1, yeast BUB1, and yeast MAD3. Identical and conserved residues in boxed region are bolded and shaded, respectively. Alignment was performed with ClustalW in MacVector 6.0.1. (c) 50 μg of HeLa lysate was probed with affinity-purified hBUBR1 antibodies (lanes 2 and 4) and affinity-purified hBUB1 antibodies (lane 1 and 3). A filter probed with hBUB1 antibodies (lane 3) was stripped and re-probed with hBUBR1 antibodies (lane 4), to show that hBUB1 and hBUBR1 can be resolved from each other. The bars show the separation of the two bands if lanes 3 and 4 were superimposed.
Figure 5
Identification of human hBUBR1 kinase. (a) Dendrogram depicting the relatedness of yeast, mouse, rat (partial clone), and human BUB1 kinases was generated with GCG PILEUP program. (b) Alignment of the NH2-terminal portion of mBUB1, hBUBR1, yeast BUB1, and yeast MAD3. Identical and conserved residues in boxed region are bolded and shaded, respectively. Alignment was performed with ClustalW in MacVector 6.0.1. (c) 50 μg of HeLa lysate was probed with affinity-purified hBUBR1 antibodies (lanes 2 and 4) and affinity-purified hBUB1 antibodies (lane 1 and 3). A filter probed with hBUB1 antibodies (lane 3) was stripped and re-probed with hBUBR1 antibodies (lane 4), to show that hBUB1 and hBUBR1 can be resolved from each other. The bars show the separation of the two bands if lanes 3 and 4 were superimposed.
Figure 5
Identification of human hBUBR1 kinase. (a) Dendrogram depicting the relatedness of yeast, mouse, rat (partial clone), and human BUB1 kinases was generated with GCG PILEUP program. (b) Alignment of the NH2-terminal portion of mBUB1, hBUBR1, yeast BUB1, and yeast MAD3. Identical and conserved residues in boxed region are bolded and shaded, respectively. Alignment was performed with ClustalW in MacVector 6.0.1. (c) 50 μg of HeLa lysate was probed with affinity-purified hBUBR1 antibodies (lanes 2 and 4) and affinity-purified hBUB1 antibodies (lane 1 and 3). A filter probed with hBUB1 antibodies (lane 3) was stripped and re-probed with hBUBR1 antibodies (lane 4), to show that hBUB1 and hBUBR1 can be resolved from each other. The bars show the separation of the two bands if lanes 3 and 4 were superimposed.
Figure 6
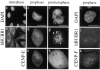
hBUBR1 assembles on kinetochores after CENP-F but before CENP-E. (b) hBUBR1 distribution in interphase HeLa cells. (c–h) Double staining of a prophase and early prometaphase cell with hBUBR1 and CENP-E. Arrowheads point to kinetochores that contain hBUBR1 (g) but not CENP-E (h). (i–k) Double staining of a prophase cell with hBUBR1 and CENP-F. In c–e and i–k, cells were extracted before fixing to reduce soluble pools of proteins that would obscure detection of kinetochore staining. hBUBR1 (b, d, g, and j) was stained with rat anti-hBUBR1 antibodies and Cy2–anti-rat IgG. CENP-E (e, h) and CENP-F (k) were stained with rabbit anti– CENP-E and CENP-F antibodies, respectively, and counterstained with Texas red–anti-rabbit IgG. Chromosomes and nuclei were stained with DAPI (a, c, f, and i). a and b were photographed with a 40× objective while other panels were photographed with a 100× objective. Bars, 10 μm.
Figure 7
hBUBR1 and CENP-E exhibit complex localization patterns during mitosis. Double-immunofluorescence staining of hBUBR1 (b, e, h, k, n) and CENP-E (c, f, i, l, o) of cells from prometaphase to telophase. The inset in d, depicts a single unaligned chromosome. Left and right arrowheads point to trailing and leading kinetochores, respectively. The trailing kinetochore exhibits stronger hBUBR1 (e) and CENP-E (f) staining than its leading kinetochore. All samples were extracted and then fixed to reduce staining contributed by the soluble pools of hBUBR1 and CENP-E. Identical antibodies were used as in Fig. 6. Bars, 10 μm.
Figure 8
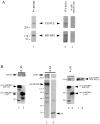
hBUBR1 forms a complex with CENP-E in HeLa cells. (a) Affinity-purified rat anti-hBUBR1 IgG was used to immunoprecipitate hBUBR1, and then probed with rabbit anti-hBUBR1 (bottom panel, lane 1) and rabbit anti–CENP-E antibodies (top panel, lane 1). Immunoprecipitates obtained with CENP-E antibodies (lane 2) and nonimmune antibodies (lane 3) were probed with CENP-E (top panel) and hBUBR1 (bottom panel). The filter was cut and the appropriate sections were probed with hBUBR1 and CENP-E antibodies. (b) ProA:hBUBR1409–1051 (lane 1), proA:hBUBR11–467 (lane 2), gfp:hBUBR1 (lane 3), and gfp (lane 4) were immunoprecipitated from transfected lysates with human IgG Sepharose and anti-gfp antibodies, respectively, and probed for coprecipitating CENP-E (lanes 1–4, top panels). Lysate prepared from cells cotransfected with gfp:CENP-E1958– 2662 and proA:hBUBR1409–1051 (lane 5) or proA:hBUBR11–429 (lane 6) were incubated with human IgG Sepharose to immunoprecipitate the proA:CENP-E fusion proteins (lanes 5 and 6, bottom panel), and then probed for coprecipitating gfp:CENP-E1958– 2662 (top panel). Expression of gfp:CENP-E1958–2662 in cells cotransfected with either proA:hBUBR1409–1051 (see lane 7) or proA:hBUBR11–467 (see lane 8) was confirmed by Western blot. ProA:CENP-E and gfp:CENP-E fusions were detected with alkaline phosphatase–conjugated human IgG and rat anti-gfp antibodies, respectively. Molecular weight standards are depicted on the left side of each of the panels.
Similar articles
- Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC.
Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Chan GK, et al. J Cell Biol. 1999 Sep 6;146(5):941-54. doi: 10.1083/jcb.146.5.941. J Cell Biol. 1999. PMID: 10477750 Free PMC article. - The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis.
Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. Jablonski SA, et al. Chromosoma. 1998 Dec;107(6-7):386-96. doi: 10.1007/s004120050322. Chromosoma. 1998. PMID: 9914370 - Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores.
Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Liu ST, et al. Mol Biol Cell. 2003 Apr;14(4):1638-51. doi: 10.1091/mbc.02-05-0074. Mol Biol Cell. 2003. PMID: 12686615 Free PMC article. - The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase.
Taylor SS, Ha E, McKeon F. Taylor SS, et al. J Cell Biol. 1998 Jul 13;142(1):1-11. doi: 10.1083/jcb.142.1.1. J Cell Biol. 1998. PMID: 9660858 Free PMC article. - Mechanisms of kinesin-7 CENP-E in kinetochore-microtubule capture and chromosome alignment during cell division.
Yu KW, Zhong N, Xiao Y, She ZY. Yu KW, et al. Biol Cell. 2019 Jun;111(6):143-160. doi: 10.1111/boc.201800082. Epub 2019 Feb 26. Biol Cell. 2019. PMID: 30784092 Review.
Cited by
- Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes.
Gui L, Homer H. Gui L, et al. Development. 2012 Jun;139(11):1941-6. doi: 10.1242/dev.078352. Epub 2012 Apr 18. Development. 2012. PMID: 22513372 Free PMC article. - BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1.
Chen RH. Chen RH. J Cell Biol. 2002 Aug 5;158(3):487-96. doi: 10.1083/jcb.200204048. Epub 2002 Aug 5. J Cell Biol. 2002. PMID: 12163471 Free PMC article. - Mitotic kinesin CENP-E promotes microtubule plus-end elongation.
Sardar HS, Luczak VG, Lopez MM, Lister BC, Gilbert SP. Sardar HS, et al. Curr Biol. 2010 Sep 28;20(18):1648-53. doi: 10.1016/j.cub.2010.08.001. Curr Biol. 2010. PMID: 20797864 Free PMC article. - CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer.
O'Brien SL, Fagan A, Fox EJ, Millikan RC, Culhane AC, Brennan DJ, McCann AH, Hegarty S, Moyna S, Duffy MJ, Higgins DG, Jirström K, Landberg G, Gallagher WM. O'Brien SL, et al. Int J Cancer. 2007 Apr 1;120(7):1434-43. doi: 10.1002/ijc.22413. Int J Cancer. 2007. PMID: 17205517 Free PMC article. - CENP-E--dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint.
Guo Y, Kim C, Ahmad S, Zhang J, Mao Y. Guo Y, et al. J Cell Biol. 2012 Jul 23;198(2):205-17. doi: 10.1083/jcb.201202152. Epub 2012 Jul 16. J Cell Biol. 2012. PMID: 22801780 Free PMC article.
References
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Wilson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. - PubMed
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials