Heterogeneous nuclear ribonucleoprotein L interacts with the 3' border of the internal ribosomal entry site of hepatitis C virus - PubMed (original) (raw)
Heterogeneous nuclear ribonucleoprotein L interacts with the 3' border of the internal ribosomal entry site of hepatitis C virus
B Hahm et al. J Virol. 1998 Nov.
Abstract
Translation initiation of hepatitis C virus (HCV) RNA occurs by internal entry of a ribosome into the 5' nontranslated region in a cap-independent manner. The HCV RNA sequence from about nucleotide 40 up to the N terminus of the coding sequence of the core protein is required for efficient internal initiation of translation, though the precise border of the HCV internal ribosomal entry site (IRES) has yet to be determined. Several cellular proteins have been proposed to direct HCV IRES-dependent translation by binding to the HCV IRES. Here we report on a novel cellular protein that specifically interacts with the 3' border of the HCV IRES in the core-coding sequence. This protein with an apparent molecular mass of 68 kDa turned out to be heterogeneous nuclear ribonucleoprotein L (hnRNP L). The binding of hnRNP L to the HCV IRES correlates with the translational efficiencies of corresponding mRNAs. This finding suggests that hnRNP L may play an important role in the translation of HCV mRNA through the IRES element.
Figures
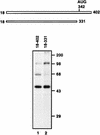
FIG. 1
Diagram of probes and detection of cellular proteins interacting with HCV IRES. RNA probes [32P]RNA(18–402) (lane 1) and [32P]RNA(18–331) (lane 2) were cross-linked to HeLa cell cytoplasmic extracts, after which the proteins were analyzed by SDS–12% PAGE. Molecular masses (in thousands) are noted to the right of the gel.
FIG. 2
Assignment of the 68-kDa protein band to hnRNP L. (A) Coprecipitation and Western blot analysis. HeLa cell extract was incubated without RNA (lane 1) or with the biotinylated HCV RNA(18–402) (lane 2) and HCV RNA(18–331) (lane 3). The bound proteins were then precipitated with streptavidin acrylamide beads. After being resolved in an SDS–12% polyacrylamide gel, hnRNP L was visualized by Western blot analysis with anti-hnRNP L monoclonal antibody. (B) Combination of RNA mobility shift and Western blot analyses. Aliquots of HeLa cell extract (20 μg) were incubated without RNA (lane 1) or with 100 ng of RNA(18–331) (lane 2), 100 ng of RNA(18–402) (lane 3), 300 ng of RNA(18–331) (lane 4), or 300 ng of RNA(18–402) (lane 5). The samples were electrophoresed in a 5% nondenaturing gel, after which the proteins were blotted onto a nitrocellulose membrane and probed by Western blotting with anti-hnRNP L monoclonal antibody. Free hnRNP L and RNA-bound hnRNP L are marked “Free” and “Bound,” respectively. (C) Coprecipitation of 35S-labeled proteins. The in vitro-translated proteins luciferase and hnRNP L are in lanes 1 and 2, respectively. These 35S-labeled proteins were incubated with biotinylated RNA(18–402) (lanes 3 and 4) and RNA(18–331) (lane 5), after which the RNA-bound proteins were precipitated with streptavidin acrylamide beads. After the samples were washed with binding buffer, the RNA-bound proteins were resolved in an SDS–12% polyacrylamide gel. The protein bands were detected by autoradiography.
FIG. 2
Assignment of the 68-kDa protein band to hnRNP L. (A) Coprecipitation and Western blot analysis. HeLa cell extract was incubated without RNA (lane 1) or with the biotinylated HCV RNA(18–402) (lane 2) and HCV RNA(18–331) (lane 3). The bound proteins were then precipitated with streptavidin acrylamide beads. After being resolved in an SDS–12% polyacrylamide gel, hnRNP L was visualized by Western blot analysis with anti-hnRNP L monoclonal antibody. (B) Combination of RNA mobility shift and Western blot analyses. Aliquots of HeLa cell extract (20 μg) were incubated without RNA (lane 1) or with 100 ng of RNA(18–331) (lane 2), 100 ng of RNA(18–402) (lane 3), 300 ng of RNA(18–331) (lane 4), or 300 ng of RNA(18–402) (lane 5). The samples were electrophoresed in a 5% nondenaturing gel, after which the proteins were blotted onto a nitrocellulose membrane and probed by Western blotting with anti-hnRNP L monoclonal antibody. Free hnRNP L and RNA-bound hnRNP L are marked “Free” and “Bound,” respectively. (C) Coprecipitation of 35S-labeled proteins. The in vitro-translated proteins luciferase and hnRNP L are in lanes 1 and 2, respectively. These 35S-labeled proteins were incubated with biotinylated RNA(18–402) (lanes 3 and 4) and RNA(18–331) (lane 5), after which the RNA-bound proteins were precipitated with streptavidin acrylamide beads. After the samples were washed with binding buffer, the RNA-bound proteins were resolved in an SDS–12% polyacrylamide gel. The protein bands were detected by autoradiography.
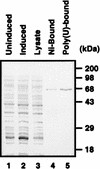
FIG. 3
Purification of hnRNP L. Purified hnRNP L was resolved by SDS–12% PAGE and stained with Coomassie brilliant blue G-250. Protein profiles in uninduced E. coli cells, in induced E. coli cells, in E. coli lysate, in E. coli cells after Ni-NTA column chromatography, and in E. coli cells after poly(U) column chromatography are shown in lanes 1, 2, 3, 4, and 5, respectively.
FIG. 4
Determination of the minimal hnRNP L-binding site in the HCV IRES. (A) UV cross-linking of hnRNP to HCV RNA. [32P]RNA(18–402) (lane 1), [32P]RNA(18–331) (lane 2), [32P]RNA(280–402) (lane 3), and [32P]RNA(331–402) (lane 4) were cross-linked to purified hnRNP L by UV irradiation. The labeled proteins were then analyzed by SDS–12% PAGE. (B) Gel mobility shift assay. RNA gel mobility shift assays were performed with [32P]RNA(228–331) (lanes 1 to 5) and [32P]RNA(342–402) (lanes 6 to 10). Samples in lanes 2 and 7 were incubated with 1 μg of BSA. Samples in lanes 3 and 8, 4 and 9, and 5 and 10 were incubated with 0.15, 0.3, and 0.6 μg of purified hnRNP L, respectively. Samples were analyzed in a 5% nondenaturing polyacrylamide gel. The positions of free RNA probe and protein-bound probe are marked “Free” and “Bound,” respectively.
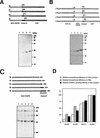
FIG. 5
Correlation between translational efficiency and hnRNP L-binding affinity. (A) Translation of monocistronic mRNAs in HeLa cell extract. Uncapped mRNAs were translated in HeLa cell extracts. Translation products were resolved by SDS–15% PAGE. Schematic diagrams of the mRNAs are shown at the top of the panel. CoreΔC, C-terminally truncated core. (B) Translation of dicistronic mRNAs in RRL. Capped dicistronic mRNAs were translated in RRL. Translation products were resolved by SDS–15% PAGE. Schematic diagrams of the mRNAs are shown at the top of the panel. CATΔC, C-terminally truncated CAT gene. (C) UV cross-linking of HCV RNAs. 32P-labeled RNA probes were cross-linked with HeLa cell extracts and then analyzed by SDS–12% PAGE. Schematic diagrams of the RNA probes are shown at the top of the panel. (D) Relative translational efficiencies of the mRNAs and relative affinities of the same set of RNAs for hnRNP L. The intensities of the bands corresponding to the polypeptides shown in panels A and B and the bands corresponding to hnRNP L in panel C were measured with a phosphorimager. The relative translational efficiencies and relative hnRNP L-binding affinities of the HCV RNAs were normalized by considering the activity of HCV RNA(18–402) 100%.
Similar articles
- hnRNP L is required for the translation mediated by HCV IRES.
Hwang B, Lim JH, Hahm B, Jang SK, Lee SW. Hwang B, et al. Biochem Biophys Res Commun. 2009 Jan 16;378(3):584-8. doi: 10.1016/j.bbrc.2008.11.091. Epub 2008 Dec 4. Biochem Biophys Res Commun. 2009. PMID: 19061868 - RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA.
Paek KY, Kim CS, Park SM, Kim JH, Jang SK. Paek KY, et al. J Virol. 2008 Dec;82(24):12082-93. doi: 10.1128/JVI.01405-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842733 Free PMC article. - Translation regulation by ribosomes: Increased complexity and expanded scope.
Mauro VP, Matsuda D. Mauro VP, et al. RNA Biol. 2016 Sep;13(9):748-55. doi: 10.1080/15476286.2015.1107701. Epub 2015 Oct 29. RNA Biol. 2016. PMID: 26513496 Free PMC article. Review. - Hepatitis C virus RNA translation.
Niepmann M. Niepmann M. Curr Top Microbiol Immunol. 2013;369:143-66. doi: 10.1007/978-3-642-27340-7_6. Curr Top Microbiol Immunol. 2013. PMID: 23463200 Review.
Cited by
- Insights into the biology of IRES elements through riboproteomic approaches.
Pacheco A, Martinez-Salas E. Pacheco A, et al. J Biomed Biotechnol. 2010;2010:458927. doi: 10.1155/2010/458927. Epub 2010 Feb 2. J Biomed Biotechnol. 2010. PMID: 20150968 Free PMC article. Review. - Internal ribosome entry sites (IRESs): reality and use.
Houdebine LM, Attal J. Houdebine LM, et al. Transgenic Res. 1999 Jun;8(3):157-77. doi: 10.1023/a:1008909908180. Transgenic Res. 1999. PMID: 10478488 Review. - The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation.
Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M. Majumder M, et al. Mol Cell Biol. 2009 May;29(10):2899-912. doi: 10.1128/MCB.01774-08. Epub 2009 Mar 9. Mol Cell Biol. 2009. PMID: 19273590 Free PMC article. - Functions of the 5' and 3' ends of calicivirus genomes.
Alhatlani B, Vashist S, Goodfellow I. Alhatlani B, et al. Virus Res. 2015 Aug 3;206:134-43. doi: 10.1016/j.virusres.2015.02.002. Epub 2015 Feb 9. Virus Res. 2015. PMID: 25678268 Free PMC article. Review. - Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes.
Brunner JE, Nguyen JH, Roehl HH, Ho TV, Swiderek KM, Semler BL. Brunner JE, et al. J Virol. 2005 Mar;79(6):3254-66. doi: 10.1128/JVI.79.6.3254-3266.2005. J Virol. 2005. PMID: 15731220 Free PMC article.
References
- Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. HnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous