Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue - PubMed (original) (raw)
Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue
J B Bowzard et al. J Virol. 1998 Nov.
Abstract
The Gag proteins of Rous sarcoma virus (RSV) and human immunodeficiency virus (HIV) contain small interaction (I) domains within their nucleocapsid (NC) sequences. These overlap the zinc finger motifs and function to provide the proper density to viral particles. There are two zinc fingers and at least two I domains within these Gag proteins. To more thoroughly characterize the important sequence features and properties of I domains, we analyzed Gag proteins that contain one or no zinc finger motifs. Chimeric proteins containing the amino-terminal half of RSV Gag and various portions of the carboxy terminus of murine leukemia virus (MLV) (containing one zinc finger) Gag had only one I domain, whereas similar chimeras with human foamy virus (HFV) (containing no zinc fingers) Gag had at least two. Mutational analysis of the MLV NC sequence and inspection of I domain sequences within the zinc-fingerless C terminus of HFV Gag suggested that clusters of basic residues, but not the zinc finger motif residues themselves, are required for the formation of particles of proper density. In support of this, a simple string of strongly basic residues was found to be able to substitute for the RSV I domains. We also explored the possibility that differences in I domains (e.g., their number) account for differences in the ability of Gag proteins to be rescued into particles when they are unable to bind to membranes. Previously published experiments have shown that such membrane-binding mutants of RSV and HIV (two I domains) can be rescued but that those of MLV (one I domain) cannot. Complementation rescue experiments with RSV-MLV chimeras now map this difference to the NC sequence of MLV. Importantly, the same RSV-MLV chimeras could be rescued by complementation when the block to budding was after, rather than before, transport to the membrane. These results suggest that MLV Gag molecules begin to interact at a much later time after synthesis than those of RSV and HIV.
Figures
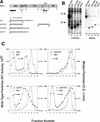
FIG. 1
The C terminus of MLV contains at least one I domain. (A) Diagram of RSV-MLV chimeras. The wild-type RSV Gag (shaded) and MLV Gag (unshaded) proteins are aligned at the junction site of the chimeras (made by fusing the _Xho_I and _Bgl_II sites of the corresponding genes). The assembly domains of RSV are indicated by black boxes and the zinc fingers in NC by hatched regions. The site of in-frame suppression of the stop codon separating the MLV gag and pol genes is marked “is.” Sites of proteolytic cleavage are indicated by vertical lines through the proteins, and numbers refer to the last amino acid of the released products. The open box and squiggled line at the N terminus of the chimeras represent replacement of the first 10 RSV amino acids with those of pp60v-src and the consequent addition of myristic acid, respectively. The MLV protease active-site mutation is indicated by a black dot. Chimera R.M1 has been previously referred to as Myr1 (37). (B) Expression of RSV-MLV chimeras. The RSV-MLV constructs were transfected into COS-1 cells, and subsequently labeled for 2.5 h with
l
-[4,5-3H(N)]leucine. The Gag proteins from the medium and lysates of the transfected cells were immunoprecipitated with a mixture of α-RSV and α-MLV antibodies, separated by SDS-PAGE, and visualized by autoradiography. The numbers to the left are the positions (in kilodaltons) of the molecular mass markers, and the position of the RSV-MLV hybrid CA cleavage product p34CA is indicated. (C) Sucrose density gradient analysis. Cells were transfected and subsequently labeled for 8 h. Medium was collected, cleared of cellular debris, mixed with purified MLV, and centrifuged for 16 h at 83,500 × g through a 10 to 50% sucrose gradient. The gradients were fractionated, and the amount of control or experimental particles in each fraction was determined by reverse transcriptase assays or immunoprecipitation followed by densitometry of autoradiograms, respectively. An arrow in each panel indicates the direction of sedimentation. O.D., optical density.
FIG. 2
An intact zinc finger motif is not required for MLV I domain activity. (A) Diagram of the chimeric BgM protein and various mutants derived from it. The precise locations of the changes made in the 60 amino acids of the MLV NC sequence are shown along with the positions of basic residues (marked with +). Substitutions are designated with arrows and deletions are boxed. The residues comprising the zinc finger motif are highlighted in gray. (B) Gag proteins were labeled and visualized and (C) sucrose gradient analysis was performed as described in the legend for Fig. 1.
FIG. 3
Basic residues can substitute for the RSV I domain. (A) p25 fusion constructs are shown with foreign amino acids denoted by their single letter code. (B) Gag proteins were labeled and visualized and (C) sucrose gradient analysis was performed as described in the legend for Fig. 1.
FIG. 4
The HFV Gag protein contains at least two I domains. (A) RSV and HFV Gag proteins are illustrated. The vertical dotted lines separate HFV Gag into regions which are analogous to the cleavage products of other retroviral Gag proteins. The black vertical bars in HFV NC indicate the position of the three GR boxes. The numbers below the chimeric proteins indicate the HFV Gag residues contained in the constructs and the letters at the end of the molecules are the single letter codes of any nonviral residues present. (B) Expression of RSV-HFV chimeras was performed as described in the legend for Fig. 1 except that
l
-[35S]methionine was used to label the cells, and the relevant proteins were immunoprecipitated with α-RSV antibody. (C) Sucrose gradient analysis was performed as described in the legend for Fig. 1 except that the RSV-HFV chimeras were labeled with
l
-[35S]methionine and immunoprecipitated with α-RSV antibody, and two constructs (Bg.2,3 and Bg.3) were mixed with labeled RSV Gag-only particles (rather than authentic MLV) before layering on the gradient.
FIG. 5
Membrane localization is required for the rescue of molecules containing the MLV I domain. (A) To assess the involvement of I domains in complementation rescue, budding-defective RSV-MLV chimeras were examined. All constructs shown here contain either two RSV I domains or the single MLV I domain. T10C.PR− and T10M.PR−, which lack an L domain, are blocked at a late step during budding, and BgM.M(−), which lacks myristate, is not targeted to the plasma membrane. Cells were transfected and labeled as described in the legend for Fig. 1. Cotransfection is denoted by brackets above the lanes and the cotransfected DNA above the bracket. (B) Unmyristylated BgM.M(−) cannot be rescued either by molecules containing RSV I domains or by the MLV I domain. However, BgM, which is targeted to the membrane, is copackaged with molecules containing either RSV or MLV I domains. (C) RSV-specific (*R) and MLV-specific (*M) cleavage products are indicated. (D) A chimera containing the MLV I domain, but no L domain (T10M), is targeted to the membrane and rescued by either RSV or MLV Gag.
Similar articles
- Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus gag proteins.
Bennett RP, Nelle TD, Wills JW. Bennett RP, et al. J Virol. 1993 Nov;67(11):6487-98. doi: 10.1128/JVI.67.11.6487-6498.1993. J Virol. 1993. PMID: 8411352 Free PMC article. - Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding.
Keller PW, Johnson MC, Vogt VM. Keller PW, et al. J Virol. 2008 Jul;82(14):6788-97. doi: 10.1128/JVI.00213-08. Epub 2008 Apr 30. J Virol. 2008. PMID: 18448521 Free PMC article. - Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the gag protein and enhance packaging and infectivity.
Mark-Danieli M, Laham N, Kenan-Eichler M, Castiel A, Melamed D, Landau M, Bouvier NM, Evans MJ, Bacharach E. Mark-Danieli M, et al. J Virol. 2005 Jun;79(12):7756-67. doi: 10.1128/JVI.79.12.7756-7767.2005. J Virol. 2005. PMID: 15919928 Free PMC article. - Properties and functions of the nucleocapsid protein in virus assembly.
Muriaux D, Darlix JL. Muriaux D, et al. RNA Biol. 2010 Nov-Dec;7(6):744-53. doi: 10.4161/rna.7.6.14065. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21157181 Free PMC article. Review. - Nucleic acid chaperone activity of retroviral Gag proteins.
Rein A. Rein A. RNA Biol. 2010 Nov-Dec;7(6):700-5. doi: 10.4161/rna.7.6.13685. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045546 Free PMC article. Review.
Cited by
- Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy.
Larson DR, Ma YM, Vogt VM, Webb WW. Larson DR, et al. J Cell Biol. 2003 Sep 29;162(7):1233-44. doi: 10.1083/jcb.200303200. J Cell Biol. 2003. PMID: 14517204 Free PMC article. - Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses.
Wang SW, Noonan K, Aldovini A. Wang SW, et al. J Virol. 2004 Jan;78(2):716-23. doi: 10.1128/jvi.78.2.716-723.2004. J Virol. 2004. PMID: 14694103 Free PMC article. - Identification and characterization of ERV-W-like sequences in Platyrrhini species provides new insights into the evolutionary history of ERV-W in primates.
Grandi N, Pisano MP, Demurtas M, Blomberg J, Magiorkinis G, Mayer J, Tramontano E. Grandi N, et al. Mob DNA. 2020 Feb 1;11:6. doi: 10.1186/s13100-020-0203-2. eCollection 2020. Mob DNA. 2020. PMID: 32021657 Free PMC article. - Structure and immunogenicity of alternative forms of the simian immunodeficiency virus gag protein expressed using Venezuelan equine encephalitis virus replicon particles.
Cecil C, West A, Collier M, Jurgens C, Madden V, Whitmore A, Johnston R, Moore DT, Swanstrom R, Davis NL. Cecil C, et al. Virology. 2007 Jun 5;362(2):362-73. doi: 10.1016/j.virol.2006.12.029. Epub 2007 Feb 1. Virology. 2007. PMID: 17275057 Free PMC article.
References
- Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. - PubMed
- Bernhard W. Electron microscopy of tumor cells and tumor viruses: a review. Cancer Res. 1958;18:491–509. - PubMed
- Bernhard W. The detection and study of tumor viruses with the electron microscope. Cancer Res. 1960;20:712–727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA47482/CA/NCI NIH HHS/United States
- R01 CA047482/CA/NCI NIH HHS/United States
- T32 CA060395/CA/NCI NIH HHS/United States
- CA60395/CA/NCI NIH HHS/United States
- R37 CA047482/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources