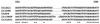Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida - PubMed (original) (raw)
Comparative Study
Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida
A Kallastu et al. J Bacteriol. 1998 Oct.
Abstract
A new insertion sequence (IS element), IS1411, was identified downstream of the phenol degradation genes pheBA that originated from plasmid DNA of Pseudomonas sp. strain EST1001. According to sequence analysis, IS1411 belongs to a new family of IS elements that has recently been named the ISL3 family (J. Mahillon and M. Chandler, Microbiol. Mol. Biol. Rev. 62:725-774, 1998). IS1411 generates 8-bp duplication of the target DNA and carries 24-bp inverted repeats (IRs), highly homologous to the IRs of other IS elements belonging to this family. IS1411 was discovered as a result of insertional activation of promoterless pheBA genes in Pseudomonas putida due to the presence of outward-directed promoters at the left end of IS1411. Both promoters located on the IS element have sequences that are similar to the consensus sequence of Escherichia coli sigma70. IS1411 can produce IS circles, and the circle formation is enhanced when two copies of the element are present in the same plasmid.
Figures
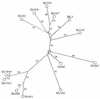
FIG. 1
Organization of the pheBA operon in plasmid pAT1140 (18). The pheB and pheA genes are flanked by two IS elements, IS_1472_ and IS_1411_ (GenBank accession no. M57500). The black boxes show the locations of the pheBA genes and the transposase genes (tnpA) of IS_1472_ and IS_1411_. The open boxes represent the intergenic regions. The promoter of the pheBA operon (designated p_i) is located upstream of IS_1472. The arrow indicates the direction of transcription of the genes. The right-end sequences of the transposon Tn_4652_ (42, 43) are shown by shaded boxes. IRR indicates the 46-bp terminal IR of the right end of Tn_4652_. (B) Organization of the pheBA operon and IS_1411_ in plasmids pEST1414 (19) and pINS113 (present study). The promoterless pheBA operon in pEST1414 is present, starting from the _Cla_I site. Only restriction sites relevant to the experiments presented in this paper are shown. C, _Cla_I; H, Hin_dIII; K, Kpn_I. The left and right IRs of IS_1411 are designated IRL and IRR, respectively. The arrow below the map of pINS113 indicates the direction of transcription of the pheBA genes from outward-directed promoters at the left end of the inserted IS_1411.
FIG. 2
Nucleotide sequence of the left end of IS_1411_. The 8-bp target sequence that was duplicated during transposition of IS_1411_ upstream of the pheBA genes is underlined with a bold line. The 24-bp IR of the element is in boldface italics. The translation start sites of pheB and tnpA of IS_1411_ are marked by double lines. The outward-directed promoters of IS_1411_ are outlined with solid lines (−10 hexamers) and dashed lines (−35 hexamers). The transcription start sites for these promoters are indicated by arrows at the coding strand of pheB. The putative −10 and −35 hexamers of the tnpA promoter are shown by solid and dashed lines, respectively. Three 5′ ends of the tnpA mRNA, mapped by reverse transcriptase, are indicated by bent arrows at the coding strand of the tnpA gene. The location of oligonucleotide (oligo) 113, used for construction of plasmid pL1411END, is indicated by the dotted arrow.
FIG. 3
Unrooted phylogenetic tree of the transposases of IS_1411_ and its related elements. Multiple alignment of transposase sequences and construction of the phylogenetic tree were carried out via the CBRG server as described in the text. PAM distances are indicated at the branches of the tree. The tree-fitting index is 0.96. DNA sequence accession numbers and hosts (in parentheses) are as follows: IS_1411_ (Pseudomonas sp.), M57500; IS_1096_ (M. smegmatis), M76495; IS_31831_ (C. glutamicum), D17429; IS_13869_ (B. lactofermentum), Z66534; IS_1396_ (Serratia marcescens), U13612; IS_1181_ (Staphylococcus aureus), L14544; IS_1193_ (Streptococcus thermophilus), Y13713; IS_1167_ (Streptococcus pneumoniae), M36180; IS_1476_ (Enterococcus faecium), U63997; IS_1165_ (Leuconostoc mesenteroides), X62617; ISL_3_ (Lactobacillus delbrueckii), X79114; IS_204_ (Nocardia asteroides), U10634; IS_1001_ (Bordetella parapertussis), X66858.
FIG. 4
Alignment of the deduced amino acid sequence of the transposase of IS_1411_ with transposases of IS_1096_, IS_13869_, and IS_31831_. Gaps introduced to optimize the alignment are shown by lines. Identical amino acids are marked by asterisks, and similar amino acids are indicated by dots. The alignments were generated via the CBRG server by using the Darwin program. Identical amino acids that were conserved in all 13 transposases analyzed in Fig. 3 are shown by shaded boxes.
FIG. 5
Sequence alignment of IRs of IS_1411_, IS_1096_, IS_204_, IS_13869_, and IS_31831_. The asterisks indicate the nucleotides conserved in all of the IS elements compared. IRR and IRL, right and left IRs, respectively.
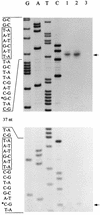
FIG. 6
Mapping of transcription initiation from outward-directed promoters of IS_1411_ by reverse transcriptase. Lanes G, A, T, and C show DNA-sequencing reactions of the left end of IS_1411_. Lanes 1 to 3 represent primer extension reactions carried out with total RNA isolated from the following bacteria: lane 1, P. putida PaW85 carrying pINS113; lane 2, E. coli HB101 carrying pINS113; lane 3, P. putida PaW85 carrying pEST1414 (negative control). The primer extension products are indicated on the right by arrows. The interrupted sequence of the left end of IS_1411_, including the −10 sequences of the promoters (boxed) and the transcription start points (indicated by asterisks), are shown on the left.
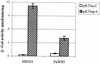
FIG. 7
β-Gal activity measured in E. coli HB101 and P. putida PaW85 carrying either the promoter-probe-vector pKTlacZ or pKTtnpA containing the tnpA promoter region. The data (means ± standard deviations) from at least four independent experiments are presented.
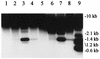
FIG. 8
IS_1411_ forms circular DNA molecules. An autoradiograph of the Southern blot of the preparations of pINS113 and pEST1414 is shown. The radioactive DNA probe was prepared from IS_1411_. Marker sizes (lane 9) are indicated on the right of the autoradiograph. Lanes: 1 and 5, pEST1414 prepared from P. putida; 2 and 6, pINS113 prepared from P. putida; 3 and 7, pINS113 prepared from E. coli; 4 and 8, pEST1414 prepared from E. coli. Lanes 1 to 4 contain uncut DNA, and lanes 5 to 8 contain DNA cut with _Kpn_I. The additional weak bands on lane 3 are of unknown origin and are not discussed in this report.
Similar articles
- Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85.
Kasak L, Hôrak R, Nurk A, Talvik K, Kivisaar M. Kasak L, et al. J Bacteriol. 1993 Dec;175(24):8038-42. doi: 10.1128/jb.175.24.8038-8042.1993. J Bacteriol. 1993. PMID: 8253692 Free PMC article. - In-vivo-generated fusion promoters in Pseudomonas putida.
Nurk A, Tamm A, Hôrak R, Kivisaar M. Nurk A, et al. Gene. 1993 May 15;127(1):23-9. doi: 10.1016/0378-1119(93)90612-7. Gene. 1993. PMID: 8387446 - Growth medium composition-determined regulatory mechanisms are superimposed on CatR-mediated transcription from the pheBA and catBCA promoters in Pseudomonas putida.
Tover A, Ojangu EL, Kivisaar M. Tover A, et al. Microbiology (Reading). 2001 Aug;147(Pt 8):2149-2156. doi: 10.1099/00221287-147-8-2149. Microbiology (Reading). 2001. PMID: 11495992 - Critical nucleotides in the interaction of CatR with the pheBA promoter: conservation of the CatR-mediated regulation mechanisms between the pheBA and catBCA operons.
Tover A, Zernant J, Chugani SA, Chakrabarty AM, Kivisaar M. Tover A, et al. Microbiology (Reading). 2000 Jan;146 ( Pt 1):173-183. doi: 10.1099/00221287-146-1-173. Microbiology (Reading). 2000. PMID: 10658664 - Transcriptional activation of the catechol and chlorocatechol operons: variations on a theme.
McFall SM, Chugani SA, Chakrabarty AM. McFall SM, et al. Gene. 1998 Nov 26;223(1-2):257-67. doi: 10.1016/s0378-1119(98)00366-7. Gene. 1998. PMID: 9858745 Review.
Cited by
- Pleiotropic Effects of Hfq on the Cytochrome c Content and Pyomelanin Production in Shewanella oneidensis.
Wang W, Liang Y, Liu L, Han S, Wu S, Gao H. Wang W, et al. Appl Environ Microbiol. 2022 Sep 22;88(18):e0128922. doi: 10.1128/aem.01289-22. Epub 2022 Sep 8. Appl Environ Microbiol. 2022. PMID: 36073941 Free PMC article. - IS2-mediated re-arrangement of the promoter sequence suppresses metabolic burden of the recombinant plasmid.
Valesová R, Stepánek V, Vecerek B, Kyslík P. Valesová R, et al. Folia Microbiol (Praha). 2005;50(4):275-82. doi: 10.1007/BF02931406. Folia Microbiol (Praha). 2005. PMID: 16408844 - Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis.
Gartemann KH, Eichenlaub R. Gartemann KH, et al. J Bacteriol. 2001 Jun;183(12):3729-36. doi: 10.1128/JB.183.12.3729-3736.2001. J Bacteriol. 2001. PMID: 11371537 Free PMC article. - Novel insertion sequence elements associated with genetic heterogeneity and phenotype conversion in Ralstonia solanacearum.
Jeong EL, Timmis JN. Jeong EL, et al. J Bacteriol. 2000 Aug;182(16):4673-6. doi: 10.1128/JB.182.16.4673-4676.2000. J Bacteriol. 2000. PMID: 10913109 Free PMC article. - How Do Transposable Elements Activate Expression of Transcriptionally Silent Antibiotic Resistance Genes?
Lipszyc A, Szuplewska M, Bartosik D. Lipszyc A, et al. Int J Mol Sci. 2022 Jul 22;23(15):8063. doi: 10.3390/ijms23158063. Int J Mol Sci. 2022. PMID: 35897639 Free PMC article. Review.
References
- Bayley S A, Duggleby C J, Worsey M J, Williams P A, Hardy K G, Broda P. Two modes of loss of the TOL function from Pseudomonas putida mt-2. Mol Gen Genet. 1977;154:203–204. - PubMed
- Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. - PubMed
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources