Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates - PubMed (original) (raw)
Comparative Study
Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates
R Patel et al. J Clin Microbiol. 1998 Nov.
Abstract
The 16S rRNA sequences of enterococcal species E. faecium, E. faecalis, E. gallinarum, E. casseliflavus/flavescens, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius are presented herein. These data were utilized to confirm the species identification of two nonmotile E. gallinarum isolates which had been previously phenotypically identified as E. faecium. The implications of this finding are discussed.
Figures
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
FIG. 1
16S rRNA sequences of E. faecium, E. faecalis, E. gallinarum, the E. casseliflavus/flavescens group, E. dispar, E. pseudoavium, E. sulfureus, E. malodoratus, E. raffinosus, E. cecorum, E. hirae, E. saccharolyticus, E. seriolicida, E. mundtii, E. avium, E. durans, E. columbae, and E. solitarius. Position 99 corresponds to position 100 of the Escherichia coli 16S rRNA gene. R = A or G; Y = C or T.
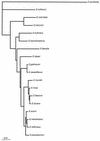
FIG. 2
Distance matrix tree of Enterococcus spp. derived from sequence homology determinations of 16S rRNA. The tree was constructed by the neighbor-joining method (23). The tree was rooted by using E. seriolicida as an outgroup. The scale bar represents a 1% difference in nucleotide sequence, as determined by taking the sum of all of the horizontal lines connecting two species.
Similar articles
- Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci.
Poyart C, Quesnes G, Trieu-Cuot P. Poyart C, et al. J Clin Microbiol. 2000 Jan;38(1):415-8. doi: 10.1128/JCM.38.1.415-418.2000. J Clin Microbiol. 2000. PMID: 10618129 Free PMC article. - Identification of enterococci at the species level by sequencing of the genes for D-alanine:D-alanine ligases.
Ozawa Y, Courvalin P, Gaiimand M. Ozawa Y, et al. Syst Appl Microbiol. 2000 Jun;23(2):230-7. doi: 10.1016/s0723-2020(00)80009-0. Syst Appl Microbiol. 2000. PMID: 10930075 - Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences.
Goh SH, Facklam RR, Chang M, Hill JE, Tyrrell GJ, Burns EC, Chan D, He C, Rahim T, Shaw C, Hemmingsen SM. Goh SH, et al. J Clin Microbiol. 2000 Nov;38(11):3953-9. doi: 10.1128/JCM.38.11.3953-3959.2000. J Clin Microbiol. 2000. PMID: 11060051 Free PMC article. - Intrageneric relationships of Enterococci as determined by reverse transcriptase sequencing of small-subunit rRNA.
Williams AM, Rodrigues UM, Collins MD. Williams AM, et al. Res Microbiol. 1991 Jan;142(1):67-74. doi: 10.1016/0923-2508(91)90098-u. Res Microbiol. 1991. PMID: 1712504 Review. - [Glycopeptide resistant enterococci. Occurrence, distribution, resistance transmission, significance].
Klare I, Witte W. Klare I, et al. Wien Klin Wochenschr. 1997 May 9;109(9):293-300. Wien Klin Wochenschr. 1997. PMID: 9265387 Review. German. No abstract available.
Cited by
- Development of a PCR assay for rapid detection of enterococci.
Ke D, Picard FJ, Martineau F, Ménard C, Roy PH, Ouellette M, Bergeron MG. Ke D, et al. J Clin Microbiol. 1999 Nov;37(11):3497-503. doi: 10.1128/JCM.37.11.3497-3503.1999. J Clin Microbiol. 1999. PMID: 10523541 Free PMC article. - Evaluation of the revised MicroScan dried overnight gram-positive identification panel to identify Enterococcus species.
Iwen PC, Rupp ME, Schreckenberger PC, Hinrichs SH. Iwen PC, et al. J Clin Microbiol. 1999 Nov;37(11):3756-8. doi: 10.1128/JCM.37.11.3756-3758.1999. J Clin Microbiol. 1999. PMID: 10523594 Free PMC article. - Diversity of domain V of 23S rRNA gene sequence in different Enterococcus species.
Tsiodras S, Gold HS, Coakley EP, Wennersten C, Moellering RC Jr, Eliopoulos GM. Tsiodras S, et al. J Clin Microbiol. 2000 Nov;38(11):3991-3. doi: 10.1128/JCM.38.11.3991-3993.2000. J Clin Microbiol. 2000. PMID: 11060057 Free PMC article. - Application of tRNA intergenic spacer PCR for identification of Enterococcus species.
Baele M, Baele P, Vaneechoutte M, Storms V, Butaye P, Devriese LA, Verschraegen G, Gillis M, Haesebrouck F. Baele M, et al. J Clin Microbiol. 2000 Nov;38(11):4201-7. doi: 10.1128/JCM.38.11.4201-4207.2000. J Clin Microbiol. 2000. PMID: 11060090 Free PMC article. - Selection of cross-resistance following exposure of Pseudomonas aeruginosa clinical isolates to ciprofloxacin or cefepime.
Alyaseen SA, Piper KE, Rouse MS, Steckelberg JM, Patel R. Alyaseen SA, et al. Antimicrob Agents Chemother. 2005 Jun;49(6):2543-5. doi: 10.1128/AAC.49.6.2543-2545.2005. Antimicrob Agents Chemother. 2005. PMID: 15917569 Free PMC article.
References
- Devriese L A, Pot B, Kersters K, Lauwers S, Haesebrouck F. Acidification of methyl-α-d-glucopyranoside: a useful test to differentiate Enterococcus casseliflavus and Enterococcus gallinarum from Enterococcus faecium species group and from Enterococcus faecalis. J Clin Microbiol. 1996;34:2607–2608. - PMC - PubMed
- Domenech A, Prieta J, Fernandez-Garayzabal J F, Collins M D, Jones D, Dominguez L. Phenotypic and phylogenetic evidence for a close relationship between Lactococcus garvieae and Enterococcus seriolicida. Microbiologia. 1993;9:63–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases