Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz - PubMed (original) (raw)
Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz
R Y Tsai et al. Mol Cell Biol. 1998 Nov.
Abstract
Roaz, a rat C2H2 zinc finger protein, plays a role in the regulation of olfactory neuronal differentiation through its interaction with the Olf-1/EBF transcription factor family. An additional role for the Roaz/Olf-1/EBF heterodimeric protein is suggested by its ability to regulate gene activation at a distinct promoter lacking Olf-1/EBF-binding sites. Using an in vitro binding-site selection assay (Selex), we demonstrate that Roaz protein binds to novel inverted perfect or imperfect repeats of GCACCC separated by 2 bp. We show that Roaz is capable of binding to a canonical consensus recognition sequence with high affinity (Kd = 3 nM). Analysis of the structural requirement for protein dimerization and DNA binding by Roaz reveals the role of specific zinc finger motifs in the Roaz protein for homodimerization and heterodimerization with the Olf-1/EBF transcription factor. The DNA-binding domain of Roaz is mapped to the N-terminal 277 amino acids, containing the first seven zinc finger motifs, which confers weak monomeric binding to a single half site and a stronger dimeric binding to the inverted repeat in a binding-site-dependent manner. Full-length protein can form dimers on both the inverted repeat and direct repeat but not on a single half site. These findings support the role of the TFIIIA-type Zn fingers in both protein-protein interaction and protein-DNA interaction and suggest distinct functions for specific motifs in proteins with a large number of zinc finger structures.
Figures
FIG. 1
Schematic diagram and sequence comparison of the 29 zinc fingers of Roaz. A schematic diagram of Roaz with individual zinc finger structures represented by shaded boxes is depicted at the top. The amino acid alignment of the 29 zinc fingers of Roaz is shown below. The zinc-coordinating Cys and His residues are shown in bold letters, and the conserved aromatic amino acids and branched aliphatic amino acids are underlined. ZF, zinc finger.
FIG. 2
Roaz-binding sites from the Selex assay. Sequence alignment of 34 isolates from the final round of PCR amplification is shown. The top 6 sequences were derived from 20 isolates from Selex for the top band; the bottom eight sequences were derived from 14 isolates for the middle band. The conserved sequences are underlined, and the two palindromic repeats are shown in bold letters. In six of the sequences, part of the consensus is contributed by the adjacent primer sequence (parentheses). The number of appearance is given in the right column. Italics indicate non-consensus primer sequence.
FIG. 3
DNA-binding affinity study. (A) A mixture of complex probes (2 pM) from the Selex for the middle band (left) or top band (right) was mixed with purified GST-Roaz, at increasing concentrations as indicated, in the EMSA. (B and C). Parallel experiments to that in panel A, except that a canonical palindromic repeat (9H5) or a direct repeat with 13 bp (X2SX, GCACCCATCGTCGAGATTAGCACCC) was used as a probe in the EMSA, respectively.
FIG. 4
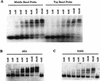
Biochemical characterization of the Roaz protein-protein interaction. (A). Purified proteins (6 to 8 μg of GST or GST-RoazD86 fusion protein bound to 20 μl of a 50% slurry of glutathione-agarose beads) were mixed with whole-cell proteins (400 μg) isolated from HEK293 cells transfected with pCIS, pCIS-XPIC2, or pCIS-XPRoazD86. Bound proteins were extracted with 25 μl of sample buffer, and a portion (8 μl) was fractionated by SDS-PAGE (10% polyacrylamide) and detected by Western blotting with anti-XPress antibody (Invitrogen). A 1/25 portion of the input sample was loaded for comparison. (B) GST fusions of RoazD86 (2 μg) (left) or OED5 (3 μg) (right) were mixed with whole-cell extract (300 μg) isolated from HEK-293 cells transfected with pCIS-XPRoazD86 pretreated with 1 mM EDTA (lane 2), 5 mM EDTA (lane 3), or 1 mM 1,10-phenanthroline (lane 4) for the in vitro binding assay. Half of the bound protein was fractionated by SDS-PAGE (10% acrylamide). A 1/40 portion of the input sample was loaded in lane 5 for comparison.
FIG. 4
Biochemical characterization of the Roaz protein-protein interaction. (A). Purified proteins (6 to 8 μg of GST or GST-RoazD86 fusion protein bound to 20 μl of a 50% slurry of glutathione-agarose beads) were mixed with whole-cell proteins (400 μg) isolated from HEK293 cells transfected with pCIS, pCIS-XPIC2, or pCIS-XPRoazD86. Bound proteins were extracted with 25 μl of sample buffer, and a portion (8 μl) was fractionated by SDS-PAGE (10% polyacrylamide) and detected by Western blotting with anti-XPress antibody (Invitrogen). A 1/25 portion of the input sample was loaded for comparison. (B) GST fusions of RoazD86 (2 μg) (left) or OED5 (3 μg) (right) were mixed with whole-cell extract (300 μg) isolated from HEK-293 cells transfected with pCIS-XPRoazD86 pretreated with 1 mM EDTA (lane 2), 5 mM EDTA (lane 3), or 1 mM 1,10-phenanthroline (lane 4) for the in vitro binding assay. Half of the bound protein was fractionated by SDS-PAGE (10% acrylamide). A 1/40 portion of the input sample was loaded in lane 5 for comparison.
FIG. 5
Regions essential for hetero- and homomultimerization of Roaz. (A). Yeast strain Y190, harboring the GAL4 DNA-binding domain fusions to Olf-1–EBF or RoazD86, was transformed with constructs encoding C-terminal deletions (cnd), N-terminal deletions (nnd), internal deletions (ind), or broken-finger mutants (bzf) of RoazD86 as GAL4 transactivator domain fusions. Double transformants were assayed for β-galactosidase activity. The positions of the amino acids (aa) which define the deletions in the nnd, cnd, and ind constructs and the zinc finger which is mutated in the bzf constructs are indicated in parentheses. The strength of interaction for an individual mutant is expressed as a percentage relative to that of intact proteins, which is set at 100% and corresponds to 43 ± 7 U for Roaz–Olf-1–EBF and 26 ± 5 U for Roaz-Roaz interactions. All measurements were determined for at least four independent colonies. A schematic diagram of Gal(TA)-RoazD86, with each shaded box representing a zinc finger structure, is shown at the top. Numbers under the boxes refer to the positions of the fingers. Mutated zinc fingers in the bzf constructs are indicated by solid ovals. Zf, zinc finger. (B) Biochemical characterization of heterodimerization between Roaz mutants and Olf-1–EBF. Whole-cell extracts (100 μg) from HEK-293 cells transfected with pCIS–Olf-1–EBF were mixed with 50 ng of individual GST or GST fusions bound to Sepharose beads as indicated. Half of the retained proteins and 2.8 μg of input whole-cell extracts were resolved by SDS-PAGE and Western blotted with either anti-Olf-1–EBF antiserum (left) or anti-GST antiserum (right). (C) Biochemical characterization of homodimerization with Roaz mutants. Whole-cell extracts (400 μg) from HEK-293 cells transfected with pCIS-XPRoazD86 were mixed with 4 μg of individual GST or GST fusions bound to Sepharose beads as indicated. One-third of the retained proteins and 10 μg of input whole-cell extracts were resolved by SDS-PAGE (10% polyacrylamide) and Western blotted with either anti-XPress antiserum (left) or anti-GST antiserum (right). Solid circles indicate the positions of aggregates of GST multimers, and asterisks indicate proteolytic products.
FIG. 6
Characterization of DNA-binding domains in Roaz. (A) Schematic diagram of C-terminally truncated Roaz proteins (CD1, CD2, CD3, and CD4) and broken-finger mutant Roaz(bzf277). Numbers in parentheses indicate the starting and ending amino acids (aa). (B) DNA sequences of synthetic oligonucleotides containing a consensus half site (X1.RBS), a consensus inverted repeat (X2.RBS), a mutated inverted repeat (X2.MUT) and a direct repeat (X2SX). The consensus sequences recognized by Roaz are indicated by bold letters. (C to E) EMSA with 32P-labeled probes as indicated and truncated forms of recombinant Roaz, full-length protein, single broken-finger mutant (bzf277), and control lysate (Ctrl) as shown in panel A. The positions of dimeric and monomeric forms of recombinant proteins are indicated by circles and asterisks, respectively. FP, free probe. (C and D) 6% polyacrylamide gel. (E) 1.5% agarose gel.
Similar articles
- EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains.
Hagman J, Gutch MJ, Lin H, Grosschedl R. Hagman J, et al. EMBO J. 1995 Jun 15;14(12):2907-16. doi: 10.1002/j.1460-2075.1995.tb07290.x. EMBO J. 1995. PMID: 7796816 Free PMC article. - Combining structure-based design with phage display to create new Cys(2)His(2) zinc finger dimers.
Wolfe SA, Ramm EI, Pabo CO. Wolfe SA, et al. Structure. 2000 Jul 15;8(7):739-50. doi: 10.1016/s0969-2126(00)00161-1. Structure. 2000. PMID: 10903945 - The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation.
Klug A. Klug A. Q Rev Biophys. 2010 Feb;43(1):1-21. doi: 10.1017/S0033583510000089. Epub 2010 May 18. Q Rev Biophys. 2010. PMID: 20478078 Review. - Zinc-finger transcription factors in plants.
Takatsuji H. Takatsuji H. Cell Mol Life Sci. 1998 Jun;54(6):582-96. doi: 10.1007/s000180050186. Cell Mol Life Sci. 1998. PMID: 9676577 Free PMC article. Review.
Cited by
- Identification of a recurrent transforming UBR5-ZNF423 fusion gene in EBV-associated nasopharyngeal carcinoma.
Chung GT, Lung RW, Hui AB, Yip KY, Woo JK, Chow C, Tong CY, Lee SD, Yuen JW, Lun SW, Tso KK, Wong N, Tsao SW, Yip TT, Busson P, Kim H, Seo JS, O'Sullivan B, Liu FF, To KF, Lo KW. Chung GT, et al. J Pathol. 2013 Oct;231(2):158-67. doi: 10.1002/path.4240. J Pathol. 2013. PMID: 23878065 Free PMC article. - Stemness-related factor Sall4 interacts with transcription factors Oct-3/4 and Sox2 and occupies Oct-Sox elements in mouse embryonic stem cells.
Tanimura N, Saito M, Ebisuya M, Nishida E, Ishikawa F. Tanimura N, et al. J Biol Chem. 2013 Feb 15;288(7):5027-38. doi: 10.1074/jbc.M112.411173. Epub 2012 Dec 26. J Biol Chem. 2013. PMID: 23269686 Free PMC article. - Strain-Dependent Modifier Genes Determine Survival in Zfp423 Mice.
Alcaraz WA, Liu Z, Valdes P, Chen E, Valdovino Gonzalez AG, Wade S, Wong C, Kim E, Chen HM, Ponn A, Concepcion D, Hamilton BA. Alcaraz WA, et al. G3 (Bethesda). 2020 Nov 5;10(11):4241-4247. doi: 10.1534/g3.120.401720. G3 (Bethesda). 2020. PMID: 32967895 Free PMC article. - Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes.
Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, Rosen ED. Griffin MJ, et al. J Biol Chem. 2013 Dec 13;288(50):35925-39. doi: 10.1074/jbc.M113.491936. Epub 2013 Oct 30. J Biol Chem. 2013. PMID: 24174531 Free PMC article. - Identification of rice Di19 family reveals OsDi19-4 involved in drought resistance.
Wang L, Yu C, Chen C, He C, Zhu Y, Huang W. Wang L, et al. Plant Cell Rep. 2014 Dec;33(12):2047-62. doi: 10.1007/s00299-014-1679-3. Epub 2014 Sep 20. Plant Cell Rep. 2014. PMID: 25236158
References
- Anthony-Cahill S J, Benfield P A, Fairman R, Wasserman Z R, Brenner S L, Stafford W D, Altenbach C, Hubbell W L, DeGrado W F. Molecular characterization of helix-loop-helix peptides. Science. 1992;255:979–983. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995.
- Brown R S, Sander C, Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985;186:271–274. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases