Identification and characterization of a family of mammalian methyl-CpG binding proteins - PubMed (original) (raw)
Identification and characterization of a family of mammalian methyl-CpG binding proteins
B Hendrich et al. Mol Cell Biol. 1998 Nov.
Abstract
Methylation at the DNA sequence 5'-CpG is required for mouse development. MeCP2 and MBD1 (formerly PCM1) are two known proteins that bind specifically to methylated DNA via a related amino acid motif and that can repress transcription. We describe here three novel human and mouse proteins (MBD2, MBD3, and MBD4) that contain the methyl-CpG binding domain. MBD2 and MBD4 bind specifically to methylated DNA in vitro. Expression of MBD2 and MBD4 tagged with green fluorescent protein in mouse cells shows that both proteins colocalize with foci of heavily methylated satellite DNA. Localization is disrupted in cells that have greatly reduced levels of CpG methylation. MBD3 does not bind methylated DNA in vivo or in vitro. MBD1, MBD2, MBD3, and MBD4 are expressed in somatic tissues, but MBD1 and MBD2 expression is reduced or absent in embryonic stem cells which are known to be deficient in MeCP1 activity. The data demonstrate that MBD2 and MBD4 bind specifically to methyl-CpG in vitro and in vivo and are therefore likely to be mediators of the biological consequences of the methylation signal.
Figures
FIG. 1
Comparison of MBD family proteins. (A) Amino acid alignment of the MBDs from murine MBD1, MBD2, MBD3, MBD4, and MeCP2 proteins. Amino acids identical in three or more proteins are indicated by shading. The location of an intron present in all five genes is indicated. (B) Comparison of MBD protein structure. Proteins are indicated as horizontal lines, and MBDs are shown as labeled white boxes. Two forms of MBD2 are shown corresponding to initiation of translation at either the first (MBD2a) or second (MBD2b) methionine codons (see text). The cysteine-rich repeats of MBD1 are represented by two black boxes, and the repression domain of MeCP2 is indicated as a stippled box. The glycine-arginine repeat in MBD2a and the C-terminal glutamic acid repeat of MBD3 are indicated as horizontal lines beneath the respective proteins. Alternate splice events are indicated as diagonal lines above or below the line of the protein. Any novel translation termination codons introduced in alternate splice events are indicated. The brain-specific splice of MBD1 and testis-specific splice of MBD2 are labeled B and T, respectively. Predicted molecular masses (in kilodaltons) and isoelectric points of each protein are also listed. The isoelectric point of the MBD3 protein is given both for the full-length protein and, in parentheses, for the protein excluding the glutamic acid repeat. aa, amino acids.
FIG. 2
Amino acid alignment of murine MBD2 and MBD3. Identical residues are indicated with a vertical line between the sequences; homologous residues are indicated with a colon, and similar residues are indicated with a period. Boxed amino acids correspond to the MBD. The glutamic acid repeat of MBD3 is represented by (E12). Amino acid numbering for MBD2 begins from the upstream initiator methionine (see MBD2a in Fig. 1B).
FIG. 3
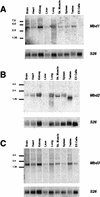
Expression of MBD1 to MBD3. cDNAs corresponding to Mbd1 (A), Mbd2 (B), and Mbd3 (C) were hybridized to Northern blots containing 10 μg of total RNA per lane from various murine tissues and ES cells. Size markers (in kilobases) are shown to the left of each blot. Blots were stripped and rehybridized to a probe corresponding to mouse ribosomal protein 26 cDNA (S26) to act as an RNA loading control. Sk. Muscle, skeletal muscle.
FIG. 4
Binding of recombinant MBD proteins to methylated DNA in vitro. One hundred picograms of radiolabeled, double-stranded GAC (lanes 1, 3, 7, 11, and 15) or GAM12 (lanes 2, 4 to 6, 8 to 10, 12 to 14, and 16 to 18) DNA (Table 1) was incubated with recombinant MBD proteins in the presence (+) or absence (−) of 200 ng of unlabeled methylated or unmethylated competitor DNA and then electrophoresed through a 2% agarose gel. The location of the free probe is indicated, and protein-DNA interactions are evidenced as slower-migrating DNA. Approximate amounts of recombinant protein used were as follows: MBD1, 20 ng; MBD2b, 10 ng; MBD3, 100 ng; MBD4, 100 ng. All proteins form a slower-migrating complex with methylated probe but not with the unmethylated probe. The shifts of MBD1, MBD2b, and MBD4 are prevented by the presence of excess, unlabeled, methylated DNA during incubation (M+ Competitor), but not by the addition of excess unmethylated DNA (M− Competitor). The full-length MBD2 protein also produces a methyl-specific shift (data not shown). The shift produced by MBD3 is competed by neither methylated nor unmethylated DNA and is therefore considered to be nonspecific.
FIG. 5
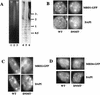
Binding of MBD proteins to methylated and hypomethylated genomes in vivo. (A) DNA from wild-type ES cells (lanes 1 and 4), DNMT mutant ES cells (lanes 2 and 5), and murine somatic cells (lanes 3 and 6) was digested with the methylation-sensitive restriction enzyme _Mae_II (Boehringer Mannheim) and electrophoresed on a 1% agarose gel. Lanes 1 to 3 show the ethidium bromide-stained gel, and lanes 4 to 6 show the gel blotted and probed with the 234-bp major satellite repeat monomer. (B to D) Localization of MBD-GFP fusion proteins in nuclei of ES cells. Either wild-type (WT) or DNMT-deficient (DNMT−) ES cells were transiently transfected with MBD1-GFP (B), MBD2-GFP (C), or MBD4-GFP (D) expression constructs. GFP fluorescence (upper panels) indicates the location of the MBD-fusion protein. DNA is visualized in the same nuclei by DAPI staining (lower panels), and major satellite is visible as intense spots of DAPI fluorescence. All fusion proteins are predominantly nuclear in all cells. MBD1-GFP colocalizes with major satellite in wild-type and mutant cells. MBD2-GFP and MBD4-GFP colocalize with major satellite in wild-type ES cells but fail to localize to major satellite in DNMT-deficient ES cell nuclei. Localization of MBD4-GFP to the nucleolus in DNMT-deficient ES cell nuclei was not observed consistently.
FIG. 6
Localization of GFP-MBD3 fusion proteins in nuclei of somatic cells. The mouse embryonic fibroblast cell line EFS2 was transiently transfected with GFP-MBD3 fusion protein expression constructs. The top panels show GFP fluorescence, while the lower panels show DAPI staining of the same cells. The constructs used are diagrammed at the bottom, with MBD depicted as a gray box and the glutamic acid repeat depicted as a black box. In all cases, GFP is located at the N terminus. (A) The full-length MBD3 fusion protein localizes to the nucleus and occasionally forms bright foci of GFP fluorescence, but these spots do not correspond to the major satellite. (B) Removal of the glutamic acid repeat (GFP-MBD3Nh) does not alter subnuclear localization, nor does removal of the N-terminal half of the MBD (GFP-MBD3Sp [C]). A fourth construct in which GFP is located at the C terminus of full-length MBD3 showed a localization pattern indistinguishable from that of GFP-MBD3 (data not shown).
Similar articles
- MBD3L1 and MBD3L2, two new proteins homologous to the methyl-CpG-binding proteins MBD2 and MBD3: characterization of MBD3L1 as a testis-specific transcriptional repressor.
Jiang CL, Jin SG, Lee DH, Lan ZJ, Xu X, O'Connor TR, Szabó PE, Mann JR, Cooney AJ, Pfeifer GP. Jiang CL, et al. Genomics. 2002 Dec;80(6):621-9. doi: 10.1006/geno.2002.7001. Genomics. 2002. PMID: 12504854 - MBD3L2 interacts with MBD3 and components of the NuRD complex and can oppose MBD2-MeCP1-mediated methylation silencing.
Jin SG, Jiang CL, Rauch T, Li H, Pfeifer GP. Jin SG, et al. J Biol Chem. 2005 Apr 1;280(13):12700-9. doi: 10.1074/jbc.M413492200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15701600 - Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes.
Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Hendrich B, et al. Mamm Genome. 1999 Sep;10(9):906-12. doi: 10.1007/s003359901112. Mamm Genome. 1999. PMID: 10441743 - Methyl-CpG-binding proteins. Targeting specific gene repression.
Ballestar E, Wolffe AP. Ballestar E, et al. Eur J Biochem. 2001 Jan;268(1):1-6. doi: 10.1046/j.1432-1327.2001.01869.x. Eur J Biochem. 2001. PMID: 11121095 Review. - Regulation of transcription and chromatin by methyl-CpG binding protein MBD1.
Nakao M, Matsui S, Yamamoto S, Okumura K, Shirakawa M, Fujita N. Nakao M, et al. Brain Dev. 2001 Dec;23 Suppl 1:S174-6. doi: 10.1016/s0387-7604(01)00348-5. Brain Dev. 2001. PMID: 11738867 Review.
Cited by
- Base-excision repair pathway shapes 5-methylcytosine deamination signatures in pan-cancer genomes.
Silveira AB, Houy A, Ganier O, Özemek B, Vanhuele S, Vincent-Salomon A, Cassoux N, Mariani P, Pierron G, Leyvraz S, Rieke D, Picca A, Bielle F, Yaspo ML, Rodrigues M, Stern MH. Silveira AB, et al. Nat Commun. 2024 Nov 14;15(1):9864. doi: 10.1038/s41467-024-54223-z. Nat Commun. 2024. PMID: 39543136 Free PMC article. - Network Analysis of Dysregulated Immune Response to COVID-19 mRNA Vaccination in Hemodialysis Patients.
Chang YS, Lee JM, Huang K, Vagts CL, Ascoli C, Edafetanure-Ibeh R, Huang Y, Cherian RA, Sarup N, Warpecha SR, Hwang S, Goel R, Turturice BA, Schott C, Martinez MH, Finn PW, Perkins DL. Chang YS, et al. Vaccines (Basel). 2024 Oct 7;12(10):1146. doi: 10.3390/vaccines12101146. Vaccines (Basel). 2024. PMID: 39460313 Free PMC article. - The therapeutic implications of all-in-one AAV-delivered epigenome-editing platform in neurodegenerative disorders.
Kantor B, O'Donovan B, Rittiner J, Hodgson D, Lindner N, Guerrero S, Dong W, Zhang A, Chiba-Falek O. Kantor B, et al. Nat Commun. 2024 Aug 23;15(1):7259. doi: 10.1038/s41467-024-50515-6. Nat Commun. 2024. PMID: 39179542 Free PMC article. - DeepPGD: A Deep Learning Model for DNA Methylation Prediction Using Temporal Convolution, BiLSTM, and Attention Mechanism.
Teragawa S, Wang L, Liu Y. Teragawa S, et al. Int J Mol Sci. 2024 Jul 26;25(15):8146. doi: 10.3390/ijms25158146. Int J Mol Sci. 2024. PMID: 39125714 Free PMC article. - Therapeutic Potential of Natural Compounds Acting through Epigenetic Mechanisms in Cardiovascular Diseases: Current Findings and Future Directions.
Bontempo P, Capasso L, De Masi L, Nebbioso A, Rigano D. Bontempo P, et al. Nutrients. 2024 Jul 24;16(15):2399. doi: 10.3390/nu16152399. Nutrients. 2024. PMID: 39125279 Free PMC article. Review.
References
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. - PubMed
- Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. - PubMed
- Bassett D E J, Boguski M S, Spencer F, Reeves R, Goebl M, Hieter P. Comparative genomics, genome cross-referencing and XREFdb. Trends Genet. 1995;11:372–373. - PubMed
- Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not embryonic cells. Genes Dev. 1995;9:2325–2334. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous