The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein - PubMed (original) (raw)
The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein
T Achsel et al. Mol Cell Biol. 1998 Nov.
Abstract
The human small nuclear ribonucleoprotein (snRNP) U5 is biochemically the most complex of the snRNP particles, containing not only the Sm core proteins but also 10 particle-specific proteins. Several of these proteins have sequence motifs which suggest that they participate in conformational changes of RNA and protein. Together, the specific proteins comprise 85% of the mass of the U5 snRNP particle. Therefore, protein-protein interactions should be highly important for both the architecture and the function of this particle. We investigated protein-protein interactions using both native and recombinant U5-specific proteins. Native U5 proteins were obtained by dissociation of U5 snRNP particles with the chaotropic salt sodium thiocyanate. A stable, RNA-free complex containing the 116-kDa EF-2 homologue (116kD), the 200kD RNA unwindase, the 220kD protein, which is the orthologue of the yeast Prp8p protein, and the U5-40kD protein was detected by sedimentation analysis of the dissociated proteins. By cDNA cloning, we show that the 40kD protein is a novel WD-40 repeat protein and is thus likely to mediate regulated protein-protein interactions. Additional biochemical analyses demonstrated that the 220kD protein binds simultaneously to the 40- and the 116kD proteins and probably also to the 200kD protein. Since the 220kD protein is also known to contact both the pre-mRNA and the U5 snRNA, it is in a position to relay the functional state of the spliceosome to the other proteins in the complex and thus modulate their activity.
Figures
FIG. 1
Dissociation of U5 snRNP particles by using sodium thiocyanate. U5 snRNPs were disrupted in 0.4 M NaSCN and separated on a 5-to-20% glycerol gradient containing the same buffer as described in Materials and Methods. Proteins were recovered from aliquots of each fraction by acetone precipitation, separated on an SDS–13% polyacrylamide gel and visualized by staining with Coomassie blue. The top 24 fractions are shown (no protein was seen in the bottom 6 fractions). The positions of the U5 proteins are indicated on the right, and those of molecular mass markers are indicated on the left. The positions to which ovalbumin (3.5S), albumin (4.6S), and immunoglobin G antibodies (7.0S) sedimented on an identical gradient are shown at the top. The bottom panel shows the RNA that was recovered by phenol extraction and ethanol precipitation, separated by urea–10% PAGE, and visualized by silver staining.
FIG. 2
Dissociated U5 snRNP particles fractionated at reduced sodium thiocyanate concentration. U5 snRNPs were disrupted in 0.4 M NaSCN as in Fig. 1 and then layered onto a 10-to-30% glycerol gradient containing 0.2 M NaSCN in the same buffer. Proteins (top panel) and RNA (bottom panel) were recovered and visualized as described for Fig. 1. The positions of the U5 proteins are indicated on the right, and those of molecular mass markers are shown on the left.
FIG. 3
The U5-200kD RNA helicase associates at 0.2 M sodium thiocyanate with the 40/116/200 complex. Fractions enriched in the 40kD, 116kD, and 220kD proteins (similar to fractions 16 and 17 in Fig. 1) or enriched in the 200kD protein (corresponding to fractions 9 to 11) were pooled from a gradient similar to that shown in Fig. 1, dialyzed separately (top and middle panels) or together (bottom panel) against buffer containing 0.2 M NaSCN, and then run on separate glycerol gradients prepared with the dialysis buffer as described in Materials and Methods. Proteins from 40 μl of 140 μl of each fraction, or from 20 μl of the sample loaded onto the gradients (first lane), were separated by SDS–10% PAGE and visualized by staining with Coomassie blue. The positions of the relevant proteins are indicated on the left.
FIG. 4
The U5-40kD protein is a novel WD-40 repeat protein. (A) Predicted amino acid sequence of the 40kD protein. Peptide sequences obtained by microsequencing are underlined; amino acids highly conserved in WD-40 repeats are shown in bold. (B) Alignment of the seven WD-40 repeats. Amino acids that agree with the consensus at the five highly conserved positions are boxed in black, and those meeting the requirements in the less conserved positions are shaded in grey. The bottom lines list all amino acids that may occur in a given position of the consensus sequence. Lowercase letters indicate groups of amino acids: h, ACMFWYVIL; t, DGNP; s, GSTACY. The brackets indicate how many nonconserved residues can be found in that region. These data are adapted from reference .
FIG. 5
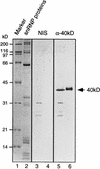
Characterization of the cDNA-encoded 40kD protein. Antibodies raised against the recombinant 40kD protein recognize the native U5 snRNP protein. Proteins present in 20 μl of nuclear extract (lanes 3 and 5) or in 5 μg of purified snRNP (lanes 4 and 6) were separated by SDS-PAGE and blotted onto nitrocellulose. The membrane was immunostained with antibodies directed against the 40kD protein (lanes 5 and 6) or preimmune serum derived from the same rabbit (lanes 3 and 4). Lanes 1 and 2 show marker proteins and snRNP proteins, respectively, which were separated on the same gel and visualized by Coomassie blue staining. Molecular mass markers are shown on the left, and the position of the 40kD protein is shown on the right.
FIG. 6
Alignment of the U5-40kD protein with highly homologous WD proteins. The U5-40kD protein was aligned with a hypothetical 38kD protein from A. thaliana (accession no. AC002333), and hypothetical proteins of 38.8 and 38.7 kDa from C. elegans (accession no. AF000265 and Z66561, respectively). The position of the WD domains is indicated by the numbered double arrows. Amino acids that are identical in at least three sequences are boxed in black, and those that are conserved in at least three sequences are shaded grey. Conserved amino acids are grouped as follows: DE, HKR, CILMVFYW, ST, and AG.
FIG. 7
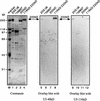
35S-labelled 40kD protein prepared by translation in vitro associates specifically with the 116/220 dimer. The translated 40kD protein was incubated in the presence of U5 snRNP particles (lanes 1 to 4) or with fractions enriched in proteins in the 1000-kDa range (lanes 5 to 8), the 200kD protein (lanes 9 to 12), or the 40kD, 116kD, and 220kD proteins (lanes 13 to 16). The protein composition of these fractions is shown in Fig. 8, left panel. Proteins precipitated with protein A-Sepharose-bound antibodies specific for the 100kD, 116kD, 200kD, or 220kD protein, as indicated above each lane, were separated by SDS-PAGE, and the 40kD protein was visualized by fluorography.
FIG. 8
The 40kD and 116kD proteins both interact with the 220kD protein on far-Western blots. U5 snRNP proteins and gradient fractions containing proteins in the 100-kDa range, the 200kD protein, or the 40kD, 116kD, and 220kD proteins were separated on three identical SDS–9% polyacrylamide gels. The proteins were either visualized by staining with Coomassie blue (left panel) or blotted onto nitrocellulose and probed with the 40kD protein prepared by translation in vitro (middle panel) or the 116kD protein (right panel) as described in Materials and Methods. The autoradiograms of the membranes are shown.
Similar articles
- The human homologue of the yeast splicing factor prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions.
Makarov EM, Makarova OV, Achsel T, Lührmann R. Makarov EM, et al. J Mol Biol. 2000 May 12;298(4):567-75. doi: 10.1006/jmbi.2000.3685. J Mol Biol. 2000. PMID: 10788320 - The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases.
Lauber J, Fabrizio P, Teigelkamp S, Lane WS, Hartmann E, Luhrmann R. Lauber J, et al. EMBO J. 1996 Aug 1;15(15):4001-15. EMBO J. 1996. PMID: 8670905 Free PMC article. - An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2.
Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. Fabrizio P, et al. EMBO J. 1997 Jul 1;16(13):4092-106. doi: 10.1093/emboj/16.13.4092. EMBO J. 1997. PMID: 9233818 Free PMC article. - CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast.
Nguyen TH, Galej WP, Fica SM, Lin PC, Newman AJ, Nagai K. Nguyen TH, et al. Curr Opin Struct Biol. 2016 Feb;36:48-57. doi: 10.1016/j.sbi.2015.12.005. Epub 2016 Jan 21. Curr Opin Struct Biol. 2016. PMID: 26803803 Free PMC article. Review. - The role of PRP8 protein in nuclear pre-mRNA splicing in yeast.
Beggs JD, Teigelkamp S, Newman AJ. Beggs JD, et al. J Cell Sci Suppl. 1995;19:101-5. doi: 10.1242/jcs.1995.supplement_19.15. J Cell Sci Suppl. 1995. PMID: 8655640 Review.
Cited by
- The U5 snRNA internal loop 1 is a platform for Brr2, Snu114 and Prp8 protein binding during U5 snRNP assembly.
Nancollis V, Ruckshanthi JP, Frazer LN, O'Keefe RT. Nancollis V, et al. J Cell Biochem. 2013 Dec;114(12):2770-84. doi: 10.1002/jcb.24625. J Cell Biochem. 2013. PMID: 23857713 Free PMC article. - Prp8 protein: at the heart of the spliceosome.
Grainger RJ, Beggs JD. Grainger RJ, et al. RNA. 2005 May;11(5):533-57. doi: 10.1261/rna.2220705. RNA. 2005. PMID: 15840809 Free PMC article. Review. - Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs.
Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Ohi MD, et al. Mol Cell Biol. 2002 Apr;22(7):2011-24. doi: 10.1128/MCB.22.7.2011-2024.2002. Mol Cell Biol. 2002. PMID: 11884590 Free PMC article. - Inhibition of SNW1 association with spliceosomal proteins promotes apoptosis in breast cancer cells.
Sato N, Maeda M, Sugiyama M, Ito S, Hyodo T, Masuda A, Tsunoda N, Kokuryo T, Hamaguchi M, Nagino M, Senga T. Sato N, et al. Cancer Med. 2015 Feb;4(2):268-77. doi: 10.1002/cam4.366. Epub 2014 Dec 1. Cancer Med. 2015. PMID: 25450007 Free PMC article. - Prp8 retinitis pigmentosa mutants cause defects in the transition between the catalytic steps of splicing.
Mayerle M, Guthrie C. Mayerle M, et al. RNA. 2016 May;22(5):793-809. doi: 10.1261/rna.055459.115. Epub 2016 Mar 11. RNA. 2016. PMID: 26968627 Free PMC article.
References
- Abramson R D, Dever T E, Merrick W C. Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J Biol Chem. 1988;263:6016–6019. - PubMed
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Asano K, Kinzy T G, Merrick W C, Hershey J W. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997;272:1101–1109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases