mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation - PubMed (original) (raw)
mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation
J M Coller et al. Genes Dev. 1998.
Abstract
Translation and mRNA stability are enhanced by the presence of a poly(A) tail. In vivo, the tail interacts with a conserved polypeptide, poly(A) binding protein (Pab1p). To examine Pab1p function in vivo, we have tethered Pab1p to the 3' UTR of reporter mRNAs by fusing it to MS2 coat protein and placing MS2 binding sites in the 3' UTR of the reporter. This strategy allows us to uncouple Pab1p function from its RNA binding activity. We show that mRNAs that lack a poly(A) tail in vivo are stabilized by Pab1p, and that the portions of Pab1p required for stabilization are genetically distinct from those required for poly(A) binding. In addition, stabilization by Pab1p requires ongoing translation of the mRNA. We conclude that the primary, or sole, function of poly(A) with respect to mRNA stability is simply to bring Pab1p to the mRNA, and that mRNA stabilization is an intrinsic property of Pab1p. The approach we describe may be useful in identifying and assaying 3' UTR regulatory proteins, as it uncouples analysis of function from RNA binding.
Figures
Figure 1
Experimental strategy. (A) The assay. To uncouple analysis of Pab1p function from the presence of a poly(A) tail, Pab1p was tethered to the 3′ UTR of reporter mRNAs by fusing it to MS2 coat protein and placing MS2-binding sites in the 3′ UTR of the reporter. For simplicity, a single-bound protein molecule is depicted; however, because MS2 coat protein binds as a dimer, at least two MS2/Pab1p molecules are likely bound per site. (B) MS2/Pab1p complements the essential functions of Pab1p in vivo. MS2/Pab1p alone complements a pab1 deletion mutation following 5-FOA selection against a plasmid bearing both the URA3 and PAB1 genes.
Figure 2
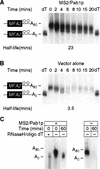
mRNA stability is conferred by MS2/Pab1p tethered to the 3′ UTR. (A) The decay of mRNAs in the presence of MS2/Pab1p, analyzed by transcriptional pulse-chase experiments and Northern blotting. The RNA and protein present in each strain is indicated above each group of six lanes, as are the orientation of MS2 sites, and the time (in min) following transcriptional repression. (Vector alone) Strains not expressing a fusion protein, but still transformed with the parental protein vector; (N) not induced sample taken prior to galactose induction. Half-lives are presented at the bottom of each panel. (B) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, shown below each lane in A. (C) RNA-binding activities of fusion proteins in yeast extracts. Gel retardation analyses were performed with crude extracts of yeast strains containing the proteins indicated at top of each group of lanes. The labeled RNA contains two MS2 coat protein recognition sites. The unlabeled competitor RNAs either contain high (wt) or low (mutant) affinity MS2 recognition sites. Mutant sites bind in vitro with an 100-fold reduction in affinity(Witherell et al. 1991). Black dot is at left of each group of three lanes indicates positions of specific complexes.
Figure 3
Tethered Pab1p stabilizes deadenylated mRNAs but does not slow poly(A) removal . A transcriptional pulse-chase experiment was performed with strains carrying MS2/Pab1p or the comparable vector. RNAs were separated through a polyacrylamide gel to resolve different lengths of poly(A) tails. Time (in min) following transcriptional repression are given above each lane. Poly(A) tail lengths of the mRNAs are shown at left; the position of the fully deadenylated RNA (A0) was determined by RNaseH/oligo (dT) cleavage of the full-length reporter. (A) RNA prepared from strain expressing MS2/Pab1p. (B) RNA prepared from strain containing the vector alone (no MS2/Pab1p). (C) Poly(A) length of mRNAs that accumulate after 60 min in the presence of MS2/Pab1p. RNA was isolated either 0 or 60 min after repression from strains with or without MS2/Pab1p, as indicated above each lane. An aliquot of RNA isolated immediately after repression (zero time) was treated with oligo(dT) and RNase H to remove poly(A) tails, and thereby provides a standard for the deadenylated reporter mRNA. Note that after 60 min, the mRNA that accumulates in the presence of MS2/Pab1p comigrates with the deadenylated mRNA standard. No mRNA is detectable in the strain lacking MS2/Pab1p after this long chase interval.
Figure 4
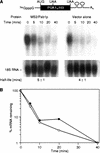
Tethered Pab1p stabilizes mRNAs that do not receive a tail. (A) Strategy. The HDV self-cleaving ribozyme was inserted into the 3′ UTR of MFA2/MS2 RNA, yielding an mRNA without a poly(A) tail (A0) in vivo. (B) Oligo(dT) cellulose chromatography. RNAs were extracted from cells carrying the ribozyme-containing construct depicted in A, or the control plasmid used in previous figures. RNAs were fractionated by oligo(dT) cellulose chromatography, then analyzed by Northern blotting. The ribozyme-containing RNA is not retained by the column, whereas the control RNA is retained. (T) Total RNA prior to fractionation; (+) retained by the column; (−) not retained by the column. The mRNA runs slightly faster in the oligo(dT) retained (+) sample than in the total (T) RNA, because less RNA is loaded per lane. (C) S1 nuclease analysis. The 3′ end of mRNA prepared from a strain carrying the ribozyme-containing RNA was determined by S1 nuclease mapping. The position of undigested probe (65 nucleotides) and of the protected species (43 nucleotides) are shown. The prominent 3′ terminus lies at the expected position for ribozyme cleavage. (D) Decay of ribozyme-cleaved RNA by a transcriptional pulse-chase experiment and Northern blotting. The turnover rate of ribozyme-cleaved reporter mRNAs was determined in strains with (left) or without (right) MS2/Pab1p. Time (in min) following transcriptional repression are given directly at top; half-lives are at bottom. (E) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, at bottom of each lane in D. (•) MS2/Pab1p; (○) vector alone.
Figure 5
Tethered Pab1p does not prevent decay through mRNA surveillance. (A) Decay of nonsense mutant-containing PGK1 mRNA in a transcriptional pulse-chase experiment and Northern blotting. Stability of a PGK1/MS2 transcript harboring a nonsense mutation at position 103 was assayed in cells expressing MS2/Pab1p (left) or vector alone (right). Time (in min) following transcriptional repression are given at top of each lane; half-lives are at bottom. (B) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, shown at bottom of each lane in A. (•) MS2/Pab1p; (○) vector alone.
Figure 6
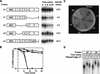
Translation is required in cis. (A) Polysome distribution of the MFA2/MS2 mRNA when bound by tethered Pab1p. The translational status of the MFA2/MS2 mRNA was determined by isolating polysomes from strains expressing either MS2/Pab1p or MS2/MS2. Sample optical trace is at top. Numbers at bottom indicate collected fractions. (B) Decay of MFA2/MS2 mRNA bearing a stem–loop in its 5′ UTR, analyzed by transcriptional pulse chase. The turnover rate of the MFA2/MS2 mRNA with or without the stem–loop in its 5′ UTR was determined in strains expressing (lanes 13–18 and lanes 19–24) or lacking (lanes 1–6 and lanes 7–12) MS2/Pab1p. Time (in min) following transcriptional repression are given at top of each lane; half-lives are presented at bottom. (C) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, shown at bottom of each lane in B. (█) Vector alone, no stem–loop; (□) vector alone, stem–loop; (•) MS2/Pab1p, no stem–loop; (○) MS2/Pab1p, stem–loop.
Figure 7
mRNA stabilization and poly(A) binding by Pab1p are genetically separable. (A) Decay of MFA2/MS2 mRNA in strains carrying MS2/Pab1p or deletion derivations, analyzed by the transcriptional pulse-chase protocol. The structure of each deletion form of MS2/Pab1p is depicted at left. Each RRM domain is designated by a number; (C) carboxy-terminal portion of Pab1p, (CΔ50) lacks the last 50 amino acids of Pab1p. Time (in min) following transcriptional repression are given at top of each lane. Half-lives are presented at right. (B) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA. (•) Wild type; (○) Δ1; (♦) Δ2; (█) Δ3; (□) Δ4. (C) Only full-length MS2/Pab1p complements the essential functions of Pab1p in vivo. The ability of plasmids carrying each form of MS2/Pab1p depicted in A to support cell growth in a strain lacking a functional PAB1 gene was determined by a plasmid shuffle assay. The assay was performed as described in Materials and Methods, and in the text (Fig. 1B). Only the wild-type construct complements a pab1 deletion following 5-FOA selection against a plasmid bearing both the URA3 and PAB1 genes. (D) Poly(A) length of mRNAs that accumulate after 8 min in the presence of Δ3 or Δ4. RNA was isolated either 0 or 8 min after repression, from strains with Δ3 or Δ4 as indicated above each lane. An aliquot of the RNA isolated immediately after repression (zero time) was treated with oligo(T) and RNase H to remove poly(A) tails, and thereby provide a standard for the deadenylated reporter mRNA.
Similar articles
- Tethering of poly(A)-binding protein interferes with non-translated mRNA decay from the 5' end in yeast.
Tsuboi T, Inada T. Tsuboi T, et al. J Biol Chem. 2010 Oct 29;285(44):33589-601. doi: 10.1074/jbc.M110.117150. Epub 2010 Aug 23. J Biol Chem. 2010. PMID: 20732870 Free PMC article. - A role for the poly(A)-binding protein Pab1p in PUF protein-mediated repression.
Chritton JJ, Wickens M. Chritton JJ, et al. J Biol Chem. 2011 Sep 23;286(38):33268-78. doi: 10.1074/jbc.M111.264572. Epub 2011 Jul 15. J Biol Chem. 2011. PMID: 21768112 Free PMC article. - The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms.
Otero LJ, Ashe MP, Sachs AB. Otero LJ, et al. EMBO J. 1999 Jun 1;18(11):3153-63. doi: 10.1093/emboj/18.11.3153. EMBO J. 1999. PMID: 10357826 Free PMC article. - Eukaryotic translation initiation: there are (at least) two sides to every story.
Sachs AB, Varani G. Sachs AB, et al. Nat Struct Biol. 2000 May;7(5):356-61. doi: 10.1038/75120. Nat Struct Biol. 2000. PMID: 10802729 Review. - Translational control of mRNAs by 3'-Untranslated region binding proteins.
Yamashita A, Takeuchi O. Yamashita A, et al. BMB Rep. 2017 Apr;50(4):194-200. doi: 10.5483/bmbrep.2017.50.4.040. BMB Rep. 2017. PMID: 28287067 Free PMC article. Review.
Cited by
- Natural and artificial binders of polyriboadenylic acid and their effect on RNA structure.
Roviello GN, Musumeci D, Roviello V, Pirtskhalava M, Egoyan A, Mirtskhulava M. Roviello GN, et al. Beilstein J Nanotechnol. 2015 Jun 17;6:1338-47. doi: 10.3762/bjnano.6.138. eCollection 2015. Beilstein J Nanotechnol. 2015. PMID: 26199837 Free PMC article. Review. - Deciphering the cellular pathway for transport of poly(A)-binding protein II.
Calado A, Kutay U, Kühn U, Wahle E, Carmo-Fonseca M. Calado A, et al. RNA. 2000 Feb;6(2):245-56. doi: 10.1017/s1355838200991908. RNA. 2000. PMID: 10688363 Free PMC article. - Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling.
Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Valentini SR, et al. Genetics. 2002 Feb;160(2):393-405. doi: 10.1093/genetics/160.2.393. Genetics. 2002. PMID: 11861547 Free PMC article. - Effect of PFOA on DNA Methylation and Alternative Splicing in Mouse Liver.
Wen Y, Chen J, Li J, Arif W, Kalsotra A, Irudayaraj J. Wen Y, et al. Toxicol Lett. 2020 Sep 1;329:38-46. doi: 10.1016/j.toxlet.2020.04.012. Epub 2020 Apr 19. Toxicol Lett. 2020. PMID: 32320774 Free PMC article. - Targeted translational regulation using the PUF protein family scaffold.
Cooke A, Prigge A, Opperman L, Wickens M. Cooke A, et al. Proc Natl Acad Sci U S A. 2011 Sep 20;108(38):15870-5. doi: 10.1073/pnas.1105151108. Epub 2011 Sep 12. Proc Natl Acad Sci U S A. 2011. PMID: 21911377 Free PMC article.
References
- Beelman C, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. - PubMed
- Beelman C, Stevens A, Caponigro G, Lagrandeur T, Hatfield L, Fortner D, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases