Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase - PubMed (original) (raw)
Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase
K Furuya et al. Genes Dev. 1998.
Abstract
The loss of sister chromatid cohesion triggers anaphase spindle movement. The budding yeast Mcd1/Scc1 protein, called cohesin, is required for associating chromatids, and proteins homologous to it exist in a variety of eukaryotes. Mcd1/Scc1 is removed from chromosomes in anaphase and degrades in G1. We show that the fission yeast protein, Mis4, which is required for equal sister chromatid separation in anaphase is a different chromatid cohesion molecule that behaves independent of cohesin and is conserved from yeast to human. Its inactivation in G1 results in cell lethality in S phase and subsequent premature sister chromatid separation. Inactivation in G2 leads to cell death in subsequent metaphase-anaphase progression but missegregation occurs only in the next round of mitosis. Mis4 is not essential for condensation, nor does it degrade in G1. Rather, it associates with chromosomes in a punctate fashion throughout the cell cycle. mis4 mutants are hypersensitive to hydroxyurea (HU) and UV irradiation but retain the ability to restrain cell cycle progression when damaged or sustaining a block to replication. The mis4 mutation results in synthetic lethality with a DNA ligase mutant. Mis4 may form a stable link between chromatids in S phase that is split rather than removed in anaphase.
Figures
Figure 1
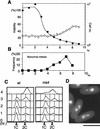
A lethal event in the first cell cycle precedes the abnormal second mitosis in a G2-synchronized culture of mis4-242 cells. Mutant cells were grown at 26°C, and small early G2 cells were collected by an elutriator rotor and then cultured at 36°C for 5 hr in synthetic EMM2 medium. (A) Cell viability (%; ▴) was measured by plating of cells at 26°C. Spindle indexes (%) were obtained by estimation of the frequencies of cells that contained the spindle stained by anti-tubulin antibody. The length of the spindle was classified into short (<2.5 μm; □) or long (>2.5 μm; █). Short spindles peaked at 80 and 200 min, whereas long spindles peaked at 100 and 220 min. The decrease of cell viability occurred when the two spindle indexes were high in the first mitosis. Abnormal mitosis occurred only in the second mitosis. (B) Mitotic cells in the first mitosis at 36°C showing normal anaphase. Micro graphs taken at 100 min. (Left) Chromatin DNA staining by DAPI; (right) spindle staining by anti-tubulin antibody. (C) Frequencies of cells showing equal sister chromatid separation in normal anaphase (○; examples shown in B) and unequal abnormal anaphase (•; examples in D). (D) Mitotic cells in the second mitosis at 36°C showing unequal sister chromatid separation. Micrographs were taken at 220 min. (Arrowheads) Chromatids remaining along the spindle. Bar, 10 μm.
Figure 2
A lethal event occurs in S phase when mis4-242 mutant cells are released from G1 arrest. Mutant cells were nitrogen source-starved at 26°C to arrest then in G1. Cells were then released in complete medium at 36°C. (A) Cell viability (•; decreasing from 3 to 4 hr after the shift) and cell number (○; increasing around 8–9 hr). (B) Frequency of cells showing unequal chromosome segregation (peaking at 7 hr). (C) DNA content measured by FACScan analysis showed that DNA replication in mis4-242 cells took place around 3–4 hr, coinciding with the occurrence of cell inviability. (wt) Wild-type control. (D) Examples of mutant cells showing abnormal anaphase. Cells stained by DAPI. Bar, 10 μm.
Figure 3
The gene product of mis4+ is similar to that of C. cinereus Rad9 and proteins present in yeast and mammals. (A) Isolation of plasmid carrying the mis4+ gene. The temperature-sensitive phenotype of mis4-242 was rescued (indicated by +) by pKT221 and some of its subclones. (Arrow) Coding region; (vertical white lines) introns. pKF201 is the minimal functional clone that contains the whole coding region. (B) Schematic representation of S. pombe Mis4, S. cerevisiae Scc2, and C. cinereus Rad9. Partial sequences derived from human and mouse cDNAs are also shown. The conserved regions (22% to 38% identity) are hatched. Mouse AA062272 and human HMHBC4244 sequences are similar to the carboxyl termini of Mis4/Scc2/Rad9. The DDBJ accession number for the nucleotide sequences of Mis4 is AB016866. (C) Immunoblot of S. pombe extracts containing Mis4–HA integrated into the chromosome (lane 2) or a plasmid containing Mis4–HA (lane 3). (lane 1) Control extracts of cells without integration and carrying the vector. (D) The immunoblot patterns of cdc25 mutant block and release experiments, showing the levels of Mis4, Cdc2 (PSTAIR), and mitotic cyclin (Cdc13). Cyclin destruction occurred 30 min after the shift to the permissive temperature (26°C). The Mis4 protein band slightly increased its mobility after 50 min (when Cdc13 levels dropped), but this was not reproducible.
Figure 4
Mis4 is sensitive to HU and becomes lethal in the S-phase block induced by HU. (A) like Cdc2-3w cells mis4-242 cells failed to form colonies in the presence of 5 m
m
HU at 26°C. (B) FACS analysis indicating that replication was blocked (DNA content is 1C) for 6 hr in mis4-242 in the presence of 15 m
m
HU at 26°C. (C) Mutant cells elongated with the interphase single nucleus. Bar, 10 μm. (D) Viability of mutant cells blocked by 15 m
m
HU remained high for 6 hr at 26°C. (E–G) Mutant cells were nitrogen starved to cause arrest in G1 at 26°C and then released in complete medium at 36°C in the presence or the absence of 15 m
m
HU. (E) The DNA content of mutant cells remained 1C for 7 hr in the presence of HU but increased to 2C after 4 hr in the absence of HU. (F) Mutant cells were elongated at 36°C in the presence of HU. (G) Viability of mutant cells decreased to 50% and 20% after 3 and 5 hr, respectively, in the absence of HU (○), whereas viability decreased to 50% and 20%, respectively, after 5 and 7 hr in the presence of HU (•). Mutant cells became inviable while the DNA content was still 1C.
Figure 5
5 UV sensitivity and instability of chromosomes in mis4-242. (A) mis4-242 cells are (•) sensitive to UV irradiation at 26°C. (○) Wild-type control. (B) The septation indices of wild-type and mis4-242 cells were equally reduced by 100 joules of UV irradiation at 26°C (•), but the recovery of the septation index to the level of normal growing cells was significantly delayed in mutant cells (○) 0 joules of UV irradiation. (C) The double mutant mis4-242–cdc17-K42 did not form colonies even at 26°C.
Figure 6
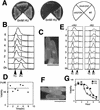
Sister chromatid separation occurs in metaphase-arrested mis4-242 cells. (A) Single cut9 and double cut9–mis4 mutant cells initially arrested in G1 by nitrogen starvation and then released in complete medium and cultured at 36°C for 8 hr. The H1 kinase activity was high in both single cut9 and double cut9–mis4 mutant cells. (B) Cut2 and Cdc13 proteolysis was absent. (C) DAPI-stained single cut9 and double cut9–mis4 mutant cells. Both mutant strains were arrested at metaphase with condensed chromosomes. (D–F) FISH analysis applied to arrested single cut9 and double cut9–mis4 cells with probes from rDNA (D) the unique centromere region cenII (E), and the unique telomere region telII (F). Counter staining by anti-SPB antibody or DAPI is also shown with the merged image. Bar, 10 μm. (G) Localization of Mis4–GFP in the nucleus. The mis4+ gene was tagged with GFP and integrated into the chromosome as a single-copy gene with the native promoter. In living cells, the GFP signal was seen in nuclear chromatin. Signals were seen in the nucleus throughout the cell cycle. Punctate nuclear signals were also seen, which appeared to localize to rDNA and telomeres. (H) Mis4–GFP signals were seen on condensed chromosomes produced in the mitotically arrested β-tubulin mutant nda3-311 at 20°C.
Figure 7
Sister centromeres are prematurely separated in the interphase state of mis4-242 after replication. The cen1 DNA was visualized by use of the LacI system; (Straight et al. 1996). Lac repressor was tagged with GFP and an NLS and expressed in a S. pombe strain containing LacO repeats integrated near cen1 (Nabeshima et al. 1998) mis4-242 cells (and a wild-type control) expressing the LacI–NLS–GFP and integrated with the LacO repeat were arrested in G1 by nitrogen starvation at 26°C, shifted to complete medium, and cultured at 36°C for 8 hr. (A) FACS analysis indicating that replication occurred in mis4-242 cells after 4 hr and was completed before 6 hr as in the wild type (not shown). (B) cen1-GFP signals were visualized, and the frequencies of cells revealing two signals (○) graphed with the frequencies of viable cells (•). Because mitotic cells started to increase after 7 hr, data for the two split signals were not taken after that time. (C) Example of mis4-242 cells showing the two split cen1 signals. The wild-type control is also shown. Bar, 10 μm.
Similar articles
- Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast.
Toyoda Y, Furuya K, Goshima G, Nagao K, Takahashi K, Yanagida M. Toyoda Y, et al. Curr Biol. 2002 Mar 5;12(5):347-58. doi: 10.1016/s0960-9822(02)00692-9. Curr Biol. 2002. PMID: 11882285 - Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase.
Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M. Tomonaga T, et al. Genes Dev. 2000 Nov 1;14(21):2757-70. doi: 10.1101/gad.832000. Genes Dev. 2000. PMID: 11069892 Free PMC article. - Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1.
Uhlmann F, Lottspeich F, Nasmyth K. Uhlmann F, et al. Nature. 1999 Jul 1;400(6739):37-42. doi: 10.1038/21831. Nature. 1999. PMID: 10403247 - Cell cycle mechanisms of sister chromatid separation; roles of Cut1/separin and Cut2/securin.
Yanagida M. Yanagida M. Genes Cells. 2000 Jan;5(1):1-8. doi: 10.1046/j.1365-2443.2000.00306.x. Genes Cells. 2000. PMID: 10651900 Review. - Secured cutting: controlling separase at the metaphase to anaphase transition.
Uhlmann F. Uhlmann F. EMBO Rep. 2001 Jun;2(6):487-92. doi: 10.1093/embo-reports/kve113. EMBO Rep. 2001. PMID: 11415980 Free PMC article. Review.
Cited by
- Replication-dependent early meiotic requirement for Spo11 and Rad50.
Merino ST, Cummings WJ, Acharya SN, Zolan ME. Merino ST, et al. Proc Natl Acad Sci U S A. 2000 Sep 12;97(19):10477-82. doi: 10.1073/pnas.190346097. Proc Natl Acad Sci U S A. 2000. PMID: 10973500 Free PMC article. - Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance.
Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Seitan VC, et al. PLoS Biol. 2006 Jul;4(8):e242. doi: 10.1371/journal.pbio.0040242. PLoS Biol. 2006. PMID: 16802858 Free PMC article. - From a single double helix to paired double helices and back.
Nasmyth K, Schleiffer A. Nasmyth K, et al. Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):99-108. doi: 10.1098/rstb.2003.1417. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065662 Free PMC article. Review. - Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1.
Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP. Feytout A, et al. Mol Cell Biol. 2011 Apr;31(8):1771-86. doi: 10.1128/MCB.01284-10. Epub 2011 Feb 7. Mol Cell Biol. 2011. PMID: 21300781 Free PMC article. - Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication.
Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Tóth A, et al. Genes Dev. 1999 Feb 1;13(3):320-33. doi: 10.1101/gad.13.3.320. Genes Dev. 1999. PMID: 9990856 Free PMC article.
References
- Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes to Cells. 1997;2:467–479. - PubMed
- Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. - PubMed
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. - PubMed
- Costello G, Rodgers L, Beach D. Fission yeast enters the stationary phase G0 state from either mitotic G1 or G2. Curr Genet. 1986;11:119–125.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases