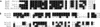Adherence of the gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins - PubMed (original) (raw)
Adherence of the gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins
R S Pegden et al. J Bacteriol. 1998 Nov.
Abstract
The adherence of Ruminococcus albus 8 to crystalline cellulose was studied, and an affinity-based assay was also used to identify candidate cellulose-binding protein(s). Bacterial adherence in cellulose-binding assays was significantly increased by the inclusion of either ruminal fluid or micromolar concentrations of both phenylacetic and phenylpropionic acids in the growth medium, and the addition of carboxymethylcellulose (CMC) to assays decreased the adherence of the bacterium to cellulose. A cellulose-binding protein with an estimated molecular mass following sodium dodecyl sulfate-polyacrylamide gel electrophoresis of approximately 21 kDa, designated CbpC, was present in both cellobiose- and cellulose-grown cultures, and the relative abundance of this protein increased in response to growth on cellulose. Addition of 0.1% (wt/vol) CMC to the binding assays had an inhibitory effect on CbpC binding to cellulose, consistent with the notion that CbpC plays a role in bacterial attachment to cellulose. The nucleotide sequence of the cbpC gene was determined by a combination of reverse genetics and genomic walking procedures. The cbpC gene encodes a protein of 169 amino acids with a calculated molecular mass of 17,655 Da. The amino-terminal third of the CbpC protein possesses the motif characteristic of the Pil family of proteins, which are most commonly involved with the formation of type 4 fimbriae and other surface-associated protein complexes in gram-negative, pathogenic bacteria. The remainder of the predicted CbpC sequence was found to have significant identity with 72- and 75-amino-acid motifs tandemly repeated in the 190-kDa surface antigen protein of Rickettsia spp., as well as one of the major capsid glycoproteins of the Chlorella virus PBCV-1. Northern blot analysis showed that phenylpropionic acid and ruminal fluid increase cbpC mRNA abundance in cellobiose-grown cells. These results suggest that CbpC is a novel cellulose-binding protein that may be involved in adherence of R. albus to substrate and extends understanding of the distribution of the Pil family of proteins in gram-positive bacteria.
Figures
FIG. 1
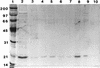
SDS-PAGE analysis of cellulose affinity (binding) assays using membrane proteins isolated from cellulose-grown cultures of R. albus 8. Lanes: 1, protein molecular mass standards; 2, aliquot of solubilized membrane proteins; 3, proteins unbound after incubation with cellulose; 4 and 5, aliquots of separate washes of the cellulose with 10 mM phosphate buffer; 6 and 7, proteins removed from cellulose by washing with 0.1% (vol/vol) Triton X-100 and 10% (wt/vol) cellobiose, respectively; 8, aliquot of SDS-PAGE running buffer mixed with the cellulose and boiled. CbpC and another low-molecular-mass cellulose-binding polypeptide are marked with arrows. Sizes are indicated in kilodaltons.
FIG. 2
SDS-PAGE analysis of cellulose affinity assay using proteins concentrated from the cell-free supernatant of cellulose-grown cultures of R. albus 8. Lanes: 1, protein molecular mass standards; 2, aliquot of concentrated extracellular proteins; 3, proteins unbound following incubation with cellulose; 4 through 7, aliquots of four separate washes of the cellulose particles with 10 mM phosphate buffer (pH 6.5); 8 and 9, proteins removed from cellulose following treatment of the cellulose particles with 0.1% (vol/vol) Triton X-100 and 10% (wt/vol) cellobiose, respectively; 10, proteins removed from cellulose particles after resuspension and boiling in SDS-PAGE running buffer. Sizes are indicated in kilodaltons.
FIG. 3
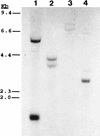
Analysis of CbpC binding to increasing amounts of cellulose, in the presence or absence of 0.1% (wt/vol) CMC. (A) Relative amount of the CbpC protein preparation bound to either 12.5 mg (lane 1), 6.25 mg (lane 2), 3.13 mg (lane 3), or 1.56 mg (lane 4) of cellulose particles as described in Material and Methods; (B) results obtained when CMC was added prior to cellulose in the assay buffer. There were no visible differences in CbpC binding to cellulose in the presence or absence of CMC when the amount of cellulose in the assay was increased to 25 mg or above.
FIG. 4
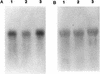
Southern blot analysis of genomic DNA isolated from R. albus 8 (lane 1), 7 (lane 2), B199 (lane 3), and SY3 (lane 4), using the 350-bp PCR product of the cbpC gene as a probe. Genomic DNA from all strains was digested with the restriction enzyme _Bgl_II, and the migration distances of standard DNA fragments are shown on the left.
FIG. 5
Nucleotide sequence of the cbpC gene from R. albus 8. The predicted amino acid sequence is shown in single-letter code above the coding sequence; putative −10, −35, and ribosome-binding sites are underlined; the stop codon is indicated by an asterisk. Sequences also determined by Edman degradation of the purified CbpC protein are double underlined, and the presumed cleavage site to remove the leader sequence is denoted by the inverted triangle.
FIG. 6
(A) Sequence alignments of the CbpC amino terminus (Ralb) with the amino-terminal sequences from the type 4 fimbrial proteins of M. bovis (Mbov), D. nodosus (Dnod), N. gonorrhoeae (Ngon), and P. aeruginosa (Paer); (B) alignment of amino acid residues 43 through 128 of CbpC with the 72- and 75-amino-acid motifs present in the surface protein antigen of R. rickettsii (Rric72 and Rric75). Identical amino acid residues are highlighted by a dark background; similar amino acid residues are indicated by the shaded background.
FIG. 7
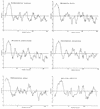
Kyte-Doolittle plots of the deduced amino acid sequences for R. albus cellulose-binding protein CbpC and type 4 fimbrial proteins from M. bovis, D. nodosus, N. gonorrhoeae, and P. aeruginosa. For the sake of comparison, the Kyte-Doolittle plot of ComG protein from B. subtilis, a nonfimbrial Pil-like protein, is included.
FIG. 8
Northern blot analysis of cbpC transcript abundance following growth in various media. Total RNA was harvested from cells following growth in cellobiose medium containing either ruminal fluid (lane 1), no additions (lane 2), or both PAA and PPA. (A) Results obtained with the _cbpC_-gene specific probe; (B) the same membrane probed with an oligonucleotide complementary to 16S rRNA.
Similar articles
- The Ruminococcus albus pilA1-pilA2 locus: expression and putative role of two adjacent pil genes in pilus formation and bacterial adhesion to cellulose.
Rakotoarivonina H, Larson MA, Morrison M, Girardeau JP, Gaillard-Martinie B, Forano E, Mosoni P. Rakotoarivonina H, et al. Microbiology (Reading). 2005 Apr;151(Pt 4):1291-9. doi: 10.1099/mic.0.27735-0. Microbiology (Reading). 2005. PMID: 15817796 - Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili.
Rakotoarivonina H, Jubelin G, Hebraud M, Gaillard-Martinie B, Forano E, Mosoni P. Rakotoarivonina H, et al. Microbiology (Reading). 2002 Jun;148(Pt 6):1871-1880. doi: 10.1099/00221287-148-6-1871. Microbiology (Reading). 2002. PMID: 12055307 - Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins?
Morrison M, Miron J. Morrison M, et al. FEMS Microbiol Lett. 2000 Apr 15;185(2):109-15. doi: 10.1111/j.1574-6968.2000.tb09047.x. FEMS Microbiol Lett. 2000. PMID: 10754233 Review. - Invited review: adhesion mechanisms of rumen cellulolytic bacteria.
Miron J, Ben-Ghedalia D, Morrison M. Miron J, et al. J Dairy Sci. 2001 Jun;84(6):1294-309. doi: 10.3168/jds.S0022-0302(01)70159-2. J Dairy Sci. 2001. PMID: 11417686 Review. - Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture.
Devillard E, Goodheart DB, Karnati SK, Bayer EA, Lamed R, Miron J, Nelson KE, Morrison M. Devillard E, et al. J Bacteriol. 2004 Jan;186(1):136-45. doi: 10.1128/JB.186.1.136-145.2004. J Bacteriol. 2004. PMID: 14679233 Free PMC article.
Cited by
- Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit.
Swierczynski A, Ton-That H. Swierczynski A, et al. J Bacteriol. 2006 Sep;188(17):6318-25. doi: 10.1128/JB.00606-06. J Bacteriol. 2006. PMID: 16923899 Free PMC article. - Are cellulosome scaffolding protein CipC and CBM3-containing protein HycP, involved in adherence of Clostridium cellulolyticum to cellulose?
Ferdinand PH, Borne R, Trotter V, Pagès S, Tardif C, Fierobe HP, Perret S. Ferdinand PH, et al. PLoS One. 2013 Jul 25;8(7):e69360. doi: 10.1371/journal.pone.0069360. Print 2013. PLoS One. 2013. PMID: 23935995 Free PMC article. - Caldicellulosiruptor bescii Adheres to Polysaccharides via a Type IV Pilin-Dependent Mechanism.
Khan AMAM, Hauk VJ, Ibrahim M, Raffel TR, Blumer-Schuette SE. Khan AMAM, et al. Appl Environ Microbiol. 2020 Apr 17;86(9):e00200-20. doi: 10.1128/AEM.00200-20. Print 2020 Apr 17. Appl Environ Microbiol. 2020. PMID: 32086304 Free PMC article. - Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis.
Christopherson MR, Dawson JA, Stevenson DM, Cunningham AC, Bramhacharya S, Weimer PJ, Kendziorski C, Suen G. Christopherson MR, et al. BMC Genomics. 2014 Dec 4;15(1):1066. doi: 10.1186/1471-2164-15-1066. BMC Genomics. 2014. PMID: 25477200 Free PMC article. - Editorial.
Filloux A. Filloux A. FEMS Microbiol Rev. 2015 Jan;39(1):1. doi: 10.1093/femsre/fuu005. FEMS Microbiol Rev. 2015. PMID: 25793962 No abstract available.
References
- Anacker R L, List R H, Mann R E, Hayes S F, Thomas L A. Characterisation of monoclonal antibodies protecting mice against Rickettsia rickettsii. J Infect Dis. 1985;151:1052–1060. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1992.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous