A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes - PubMed (original) (raw)
A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes
I Möller et al. Proc Natl Acad Sci U S A. 1998.
Abstract
For proteins to enter the secretory pathway, the membrane attachment site (M-site) on ribosomes must bind cotranslationally to the Sec61 complex present in the endoplasmic reticulum membrane. The signal recognition particle (SRP) and its receptor (SR) are required for targeting, and the nascent polypeptide associated complex (NAC) prevents inappropriate targeting of nonsecretory nascent chains. In the absence of NAC, any ribosome, regardless of the polypeptide being synthesized, binds to the endoplasmic reticulum membrane, and even nonsecretory proteins are translocated across the endoplasmic reticulum membrane. By occupying the M-site, NAC prevents all ribosome binding unless a signal peptide and SRP are present. The mechanism by which SRP overcomes the NAC block is unknown. We show that signal peptide-bound SRP occupies the M-site and therefore keeps it free of NAC. To expose the M-site and permit ribosome binding, SR can pull SRP away from the M-site without prior release of SRP from the signal peptide.
Figures
Figure 1
NAC prevents targeting by blocking the interaction of ribosomes with the Sec61 complex. High salt-stripped 77aaffLuc RNCs were prepared in a reticulocyte lysate translation system supplemented with [35]S-Met. RNCs at the final concentration of 3 OD260/ml were incubated with 1.5 μM NAC or NAC buffer for 2 min at 26°C and 5 min on ice before the addition of 1 equivalent (eq) of puromycin/KOAc-washed microsomes (PKRMs) (30) or reconstituted Sec61 complex containing proteoliposomes (17) as indicated. Total assay volumes were 20 μl. Binding was assessed with the flotation assay. Bound RNCs are recovered in top (T) fractions whereas free RNCs are in bottom (B) fractions. In vitro transcription and translation of truncated mRNAs were as described (12) for 20 min at 26°C. Salt stripping and sedimentation of RNCs was as described (5). The complexes were resuspended in blank buffer lacking nucleotides and energy-generating systems unless otherwise indicated. RNC binding was assayed as described (14). The content of radiolabeled nascent chains in each fraction was analyzed by SDS/PAGE and fluorography or scintillation counting.
Figure 2
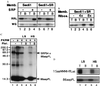
SRP blocks ribosome binding in the absence of the SR. (a) In the absence of SR, SRP blocks RNC binding to the Sec61 complex (lanes 1–4). The presence of the SR allows binding (lanes 5–8). Bound RNCs are recovered in top (T) fractions whereas free RNCs are in bottom (B) fractions. High salt-stripped 86aapPL RNCs were prepared in the reticulocyte or wheat germ lysate translation systems as described in the legend to Fig. 1. RNCs at the final concentration of 3 OD260/ml were incubated with 20 nM SRP or buffer as indicated in the figure for 5 min at 26°C and 5 min on ice before the addition of 1 eq reconstituted proteoliposomes containing either the Sec61 complex alone (lanes 1–4) or the Sec61 complex and the SR (lanes 5–8). Total assay volumes were 20 μl. After a second round of incubation as above, samples were fractionated with the RNC binding assay, and the fractions were analyzed by fluorography after SDS/PAGE. (b) Nontranslating 80 S ribosomes competitively inhibit the membrane association of 86aapPL RNCs that had been preincubated with SRP with Sec61 complex/SR proteoliposomes, indicating that RNC association occurs via interaction with the Sec61 complex rather than by the association of SRP with its receptor. Reticulocyte lysate-stripped 86aapPL RNCs were incubated with SRP as above and then were added to mixtures containing reticulocyte 80 S ribosomes at the indicated molar excesses over the RNCs and 1 eq of Sec61 complex/SR proteoliposomes in a 20-μl assay. After incubation for 3 min at 26°C and 5 min on ice, samples were analyzed for RNC binding. (c) 86aapPL RNCs containing photocrosslinker-modified lysines were prepared (16) and isolated under low salt conditions (5) before the addition of SRP to 20 nM. After 5 min at 26°C and 5 min on ice, the RNCs were irradiated, and an aliquot was analyzed (lane 1). PKRMs (1 eq) were incubated with the remainder of the RNCs for 5 min at 26°C and 5 min on ice (lane 2) in a 20-μl assay. The sample shown in lanes 3 and 4 was prepared like the one shown in lane 2 except that it was analyzed for RNC binding before SDS/PAGE and fluorography. Bound RNCs as well as the SRP 54-crosslinked nascent chains were recovered in the top fractions (lane 3). Because the RNCs containing irreversibly crosslinked SRP54 bound to the membranes, SR can free the M-site of SRP without prior release of SRP. The sample shown in lanes 5 and 6 was prepared the same as that in lanes 3 and 4, but ribosome binding was assessed under high salt conditions (0.5 M KOAc). (d) High salt-resistant binding of 15-aa-long nascent chains. The 15-aa stripped RNCs were produced by in vitro translation in a reticulocyte lysate of a truncated mRNA in which four codons encoding Met-Met-Met-Ile were engineered upstream of the sequence of the first 11 amino acids of firefly luciferase (15aaMMMI-ffLuc). 15aaMMMI-ffLuc or 86aapPL RNCs derived from 4 μl of translation reaction were incubated with 2 eq PKRMs, and, after incubation, samples were adjusted to either low (150 mM KOAc) or high salt (500 mM KOAc). Samples were fractionated in the flotation assay, in which gradients also were adjusted to 150 mM (LS) or 500 mM (HS) KOAc. Note that the short nascent chains bind as well as the longer chains (which are known to be membrane inserted) and that the fraction that binds in a salt-resistant manner is similar.
Figure 3
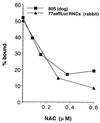
NAC prevents targeting of nontranslating ribosomes. NAC blocks the association of 80S ribosomes with ER membranes. Ribosomes prepared by puromycin and high salt treatment of dog pancreas microsomes were radiolabeled as described (18). Saturating concentrations of ribosomes (4 OD260/ml) were incubated with purified NAC at the concentrations indicated for 2 min at 26°C and 5 min on ice before the addition of 8 eq of EDTA/KOAc-washed microsomes (30, 31) in a 20-μl assay followed by another round of incubation. Binding was assessed by using the RNC binding assay described in Fig. 1. The ability of NAC to inhibit the binding of a similar amount of high salt-stripped 77aaffLuc RNCs was assayed in parallel.
Figure 4
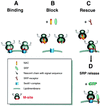
Model. (a) Ribosomes have an intrinsic ability to bind to the Sec61 complex via the M-site. In the absence of NAC and SRP, specificity is lost and any RNC can bind to the translocon. (b) NAC and SRP compete for binding to the M-site with the characteristics of the nascent chain in the vicinity of the M-site, determining which factor binds. NAC binds unless a signal peptide emerges to which SRP binds with high affinity. By experimentally “removing” SR, an intermediate where SRP blocks the M-site was detected. (c) If in the M-site, SRP first engages its receptor and then SR clears the M-site so that the ribosome can engage Sec61 complex. This can occur before the GTP-mediated release of SRP from the signal peptide (d).
Similar articles
- The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex.
Lauring B, Kreibich G, Weidmann M. Lauring B, et al. Proc Natl Acad Sci U S A. 1995 Oct 10;92(21):9435-9. doi: 10.1073/pnas.92.21.9435. Proc Natl Acad Sci U S A. 1995. PMID: 7568149 Free PMC article. - Unregulated exposure of the ribosomal M-site caused by NAC depletion results in delivery of non-secretory polypeptides to the Sec61 complex.
Möller I, Beatrix B, Kreibich G, Sakai H, Lauring B, Wiedmann M. Möller I, et al. FEBS Lett. 1998 Dec 11;441(1):1-5. doi: 10.1016/s0014-5793(98)01440-9. FEBS Lett. 1998. PMID: 9877153 - Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction.
Neuhof A, Rolls MM, Jungnickel B, Kalies KU, Rapoport TA. Neuhof A, et al. Mol Biol Cell. 1998 Jan;9(1):103-15. doi: 10.1091/mbc.9.1.103. Mol Biol Cell. 1998. PMID: 9436994 Free PMC article. - Structure, function and evolution of the signal recognition particle.
Nagai K, Oubridge C, Kuglstatter A, Menichelli E, Isel C, Jovine L. Nagai K, et al. EMBO J. 2003 Jul 15;22(14):3479-85. doi: 10.1093/emboj/cdg337. EMBO J. 2003. PMID: 12853463 Free PMC article. Review. - The Sec translocon mediated protein transport in prokaryotes and eukaryotes.
Denks K, Vogt A, Sachelaru I, Petriman NA, Kudva R, Koch HG. Denks K, et al. Mol Membr Biol. 2014 Mar-May;31(2-3):58-84. doi: 10.3109/09687688.2014.907455. Mol Membr Biol. 2014. PMID: 24762201 Review.
Cited by
- Dual recognition of the ribosome and the signal recognition particle by the SRP receptor during protein targeting to the endoplasmic reticulum.
Mandon EC, Jiang Y, Gilmore R. Mandon EC, et al. J Cell Biol. 2003 Aug 18;162(4):575-85. doi: 10.1083/jcb.200303143. Epub 2003 Aug 11. J Cell Biol. 2003. PMID: 12913112 Free PMC article. - The Role of Secretory Pathways in Candida albicans Pathogenesis.
Rollenhagen C, Mamtani S, Ma D, Dixit R, Eszterhas S, Lee SA. Rollenhagen C, et al. J Fungi (Basel). 2020 Feb 24;6(1):26. doi: 10.3390/jof6010026. J Fungi (Basel). 2020. PMID: 32102426 Free PMC article. Review. - Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria.
Fünfschilling U, Rospert S. Fünfschilling U, et al. Mol Biol Cell. 1999 Oct;10(10):3289-99. doi: 10.1091/mbc.10.10.3289. Mol Biol Cell. 1999. PMID: 10512867 Free PMC article. - Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes.
Tyedmers J, Lerner M, Bies C, Dudek J, Skowronek MH, Haas IG, Heim N, Nastainczyk W, Volkmer J, Zimmermann R. Tyedmers J, et al. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7214-9. doi: 10.1073/pnas.97.13.7214. Proc Natl Acad Sci U S A. 2000. PMID: 10860986 Free PMC article. - ATM-associated signalling triggers the unfolded protein response and cell death in response to stress.
Hotokezaka Y, Katayama I, Nakamura T. Hotokezaka Y, et al. Commun Biol. 2020 Jul 14;3(1):378. doi: 10.1038/s42003-020-1102-2. Commun Biol. 2020. PMID: 32665601 Free PMC article.
References
- Borgese N, Mok W, Kreibich G, Sabatini D D. J Mol Biol. 1974;88:559–580. - PubMed
- Walter P, Johnson A E. Annu Rev Cell Biol. 1994;10:87–119. - PubMed
- Wiedmann B, Sakai H, Davis T A, Wiedmann M. Nature (London) 1994;370:434–440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials