Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli - PubMed (original) (raw)
Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli
C V Popescu et al. Proc Natl Acad Sci U S A. 1998.
Abstract
The global regulator FNR (for fumarate nitrate reduction) controls the transcription of >100 genes whose products facilitate adaptation of Escherichia coli to growth under O2-limiting conditions. Previous Mössbauer studies have shown that anaerobically purified FNR contains a [4Fe-4S]2+ cluster that, on exposure to oxygen, is converted into a [2Fe-2S]2+ cluster, a process that decreases DNA binding by FNR. Using 57Fe Mössbauer spectroscopy of E. coli cells containing overexpressed FNR, we show here that the same cluster conversion also occurs in vivo on exposure to O2. Furthermore, the data show that a significant amount of the [4Fe-4S]2+ cluster is regenerated when the cells are shifted back to an anaerobic environment. The present study also demonstrates that 57Fe Mössbauer spectroscopy can be employed to study the in vivo behavior of (overexpressed) proteins. The use of this technique to study other iron-containing cell components is discussed.
Figures
Figure 1
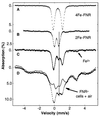
Mössbauer spectra at 4.2 K of purified 4Fe-FNR (A), of 2Fe-FNR (B), and of E. coli whole cells containing overexpressed FNR (FNR+ sample), exposed for 15 min to air (D, dash marks). Spectra of A and B were adapted from figure 1 of ref. . The solid line in D is a “background” spectrum (mostly because of ferritin) obtained by subtracting the doublet of A from the anaerobic FNR+ sample (sample of Fig. 2_B_, spectrum not shown), assuming that 4Fe-FNR represents 14% of total Fe. The difference spectrum (C, dash marks), which represents the FNR species in the air-exposed cells, was obtained by subtracting the “background” spectrum (D, solid line) from that of the air-exposed sample (D, dash marks). The solid line in C is a theoretical curve obtained by adding the doublets of 4Fe-FNR and 2Fe-FNR according to a 1:1 cluster ratio, i.e., 2:1 in Fe. The arrow indicates the Fe2+ released during cluster conversion; the vertical dotted line marks the high-energy line of the [2Fe-2S]2+ cluster. The spectra shown in D are from the same samples as those of Fig. 2_C_.
Figure 2
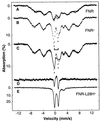
Mössbauer spectra of E. coli cells recorded at 1.5 K (A–C) and 4.2 K (E). (A) FNR− cells, anaerobic (dash marks) and after exposure to air (solid line). (B) Anaerobic FNR+ cells. Bracket marks 4Fe-FNR. (C) Anaerobic FNR+ cells from B (dash marks) and FNR+ cells exposed for 15 min to air (solid line). (D) Difference spectrum of the anaerobic FNR+ sample minus the spectrum of the air-exposed FNR+ sample, by using the spectra shown in C. The solid line in D is a theoretical difference spectrum assuming 4Fe-FNR and 2Fe-FNR in 1:1 cluster ratio. (E) Whole-cell spectra of FNR-L28H mutant, anaerobic (dash marks), and exposed to air for 20 min (solid line).
Figure 3
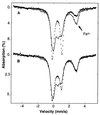
Mössbauer spectra at 4.2 K of FNR+ cells (batch different from that in Figs. 1 and 2). Spectra of anaerobic cells (A, dash marks), air-exposed cells (A and B, solid line) and cells after removal of air and incubation under argon at 4°C (B, dash marks). The arrow marks the high-energy absorption feature of Fe2+, released by cluster conversion.
Similar articles
- Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity.
Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley PJ. Khoroshilova N, et al. Proc Natl Acad Sci U S A. 1997 Jun 10;94(12):6087-92. doi: 10.1073/pnas.94.12.6087. Proc Natl Acad Sci U S A. 1997. PMID: 9177174 Free PMC article. - Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster.
Kiley PJ, Beinert H. Kiley PJ, et al. FEMS Microbiol Rev. 1998 Dec;22(5):341-52. doi: 10.1111/j.1574-6976.1998.tb00375.x. FEMS Microbiol Rev. 1998. PMID: 9990723 Review. - Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the 4Fe-4S cluster to oxygen.
Bates DM, Popescu CV, Khoroshilova N, Vogt K, Beinert H, Münck E, Kiley PJ. Bates DM, et al. J Biol Chem. 2000 Mar 3;275(9):6234-40. doi: 10.1074/jbc.275.9.6234. J Biol Chem. 2000. PMID: 10692418 - Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli.
Sutton VR, Stubna A, Patschkowski T, Münck E, Beinert H, Kiley PJ. Sutton VR, et al. Biochemistry. 2004 Jan 27;43(3):791-8. doi: 10.1021/bi0357053. Biochemistry. 2004. PMID: 14730984 - The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions.
Unden G, Schirawski J. Unden G, et al. Mol Microbiol. 1997 Jul;25(2):205-10. doi: 10.1046/j.1365-2958.1997.4731841.x. Mol Microbiol. 1997. PMID: 9282732 Review.
Cited by
- Adenosyl radical: reagent and catalyst in enzyme reactions.
Marsh EN, Patterson DP, Li L. Marsh EN, et al. Chembiochem. 2010 Mar 22;11(5):604-21. doi: 10.1002/cbic.200900777. Chembiochem. 2010. PMID: 20191656 Free PMC article. Review. - Identification and Unusual Properties of the Master Regulator FNR in the Extreme Acidophile Acidithiobacillus ferrooxidans.
Osorio H, Mettert E, Kiley P, Dopson M, Jedlicki E, Holmes DS. Osorio H, et al. Front Microbiol. 2019 Jul 19;10:1642. doi: 10.3389/fmicb.2019.01642. eCollection 2019. Front Microbiol. 2019. PMID: 31379789 Free PMC article. - Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR.
Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, Brunold TC, Markley JL, Münck E, Kiley PJ. Fleischhacker AS, et al. Biochemistry. 2012 Jun 5;51(22):4453-62. doi: 10.1021/bi3003204. Epub 2012 May 24. Biochemistry. 2012. PMID: 22583201 Free PMC article. - Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein.
Zhang B, Crack JC, Subramanian S, Green J, Thomson AJ, Le Brun NE, Johnson MK. Zhang B, et al. Proc Natl Acad Sci U S A. 2012 Sep 25;109(39):15734-9. doi: 10.1073/pnas.1208787109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23019358 Free PMC article. - Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide.
Green J, Rolfe MD, Smith LJ. Green J, et al. Virulence. 2014;5(8):794-809. doi: 10.4161/viru.27794. Epub 2014 Oct 31. Virulence. 2014. PMID: 25603427 Free PMC article. Review.
References
- Beinert H, Holm R H, Münck E. Science. 1997;277:653–659. - PubMed
- Johnson M K. Curr Opin Chem Biol. 1998;2:173–181. - PubMed
- Guest J R, Green J, Irvine A S, Spiro S. In: Regulation of Gene Expression in Escherichia coli. Lin E C C, Lynch A S, editors. Austin, TX: Landes; 1996. pp. 317–342.
- Lazazzera B A, Beinert H, Khoroshilova N, Kennedy M C, Kiley P J. J Biol Chem. 1996;271:2762–2768. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources