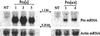Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA - PubMed (original) (raw)
Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA
P M Waterhouse et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Many examples of extreme virus resistance and posttranscriptional gene silencing of endogenous or reporter genes have been described in transgenic plants containing sense or antisense transgenes. In these cases of either cosuppression or antisense suppression, there appears to be induction of a surveillance system within the plant that specifically degrades both the transgene and target RNAs. We show that transforming plants with virus or reporter gene constructs that produce RNAs capable of duplex formation confer virus immunity or gene silencing on the plants. This was accomplished by using transcripts from one sense gene and one antisense gene colocated in the plant genome, a single transcript that has self-complementarity, or sense and antisense transcripts from genes brought together by crossing. A model is presented that is consistent with our data and those of other workers, describing the processes of induction and execution of posttranscriptional gene silencing.
Figures
Figure 1
Gene constructs. (A) Gene constructs Pro[s], Pro[a/s], and Pro[s]-stop. Pro[s] contains the Pro ORF in a sense orientation and is capable of expressing Pro protein, Pro[a/s] contains the Pro ORF in an antisense orientation, and Pro[s]-stop is the same as Pro[s] except that a thymidine residue has been inserted, making a stop codon (at the fourth codon) and a frameshift. (B) Constructs designed to express both Pro[s] and Pro[a/s] mRNAs. The Pro[s] is controlled in all constructs by the cauliflower mosaic virus 35S promoter; the terminators of Pro[s] and the promoters controlling the Pro[a/s] gene are shown. OCS, octopine synthase; term., terminator. (C) Constructs used to express ΔGUS mRNA in rice. The ΔGUS ORF has a 231-base deletion to prevent production of active GUS protein. With the exception of Gus[i/r] (i/r, inverted repeat), which is promoterless, the constructs are controlled by the ubiquitin promoter. UbiΔGus[s] and UbiΔGus[a/s] contain the ΔGUS ORFs in a sense and an antisense orientation, respectively. The 3′ region of the transcription unit of UbiΔGus[i/r] and ΔGus[i/r] is complementary to the 5′ region of the ΔGUS transcript. The predicted hairpin structure of such an mRNA is shown at the bottom.
Figure 2
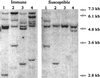
Southern blot analysis of _Hin_dIII-digested DNA (15 μg per lane) extracted from four PVY-susceptible and four PVY-immune tobacco plants containing Pro[s]-stop transgenes. The filters were hybridized with a 32P-labeled probe produced from a gel-purified DNA fragment of the 35S promoter. Each band represents one gene copy and locus.
Figure 3
Pro transgene mRNA expression in Pro[s] and Pro[a/s] plants. (Left) Total RNA from nontransgenic (NT), Pro[s]1, Pro[s]3, and Pro[s]5 tobacco plants probed for Pro sense mRNA. (Right) Nontransgenic (NT), Pro[a/s]1, and Pro[a/s]4 tobacco plants probed for antisense Pro mRNA. Both filters were also probed for actin mRNA (see Materials and Methods) for details.
Figure 4
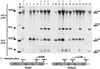
Southern blot analysis of _Apa_I-digested genomic DNA (15 μg per lane) from 20 randomly selected progeny plants (lanes 1–20) from a Pro[s]1 × Pro[a/s]1 cross. Lanes marked with “I” or unmarked contained extracts from plants subsequently shown to be immune or susceptible to PVY infection, respectively. Lane M, _M_r standard. The top portion of the filter was probed with a neomycin phosphotransferase (NPTII) sequence-specific probe to show transgene copy/locus number. The bottom portion was probed with an octopine synthase (OCS) terminator sequence-specific probe to show the presence or absence of Pro[s] and/or Pro[a/s] transgenes.
Figure 5
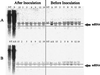
Northern blot analysis of PVY immune plants generated by crossing Pro[s]1 with Pro[a/s]4. Total RNA was extracted from nontransgenic tobacco plants (NT), immune progeny plants 2, 5, 8, 9, 12, 15, and 19, and susceptible parent plants Pro[s]1 (S) and Pro[a/s]1 (A/S) before PVY inoculation (right) and 14 days after inoculation (left). The RNAs were run on 0.9% agarose gels, transferred to membranes, and probed for Pro sense mRNA (A) and Pro antisense mRNA (B). Equal loading of samples is apparent by the similar levels of ribosomal trapping of the probe above the mRNA bands.
Figure 6
Analysis of GUS expression of supertransformed rice callus. Transgenic rice tissue containing a single Gus transgene supertransformed with UbiΔGus[s], UbiΔGus[a/s], UbiΔGus[i/r], ΔGus[i/r], and the binary vector containing the bar gene but not the ΔGus gene. For more details see Materials and Methods and Fig. 1.
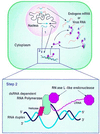
Figure 7
A model for dsRNA-induced PTGS. The transgenes transcribe both sense and antisense mRNA (step 1), which are exported to the cytoplasm where they hybridize to form a duplex. The duplex is recognized by a complex of dsRNA-dependent RNA polymerase, associated helicase, and RNase L-like molecules (step 2). The complex transcribes cRNA, attaches to it RNase L-like molecules, and releases the cRNA. The cRNA-RNase molecules hybridize to the target endogene or virus RNAs (step 3) and cleave the single-stranded regions adjacent to the hybrids (step 4), which are then degraded by other plant nucleases. This silences the expression of the endogene or provides virus immunity. The remaining RNA duplex is resistant to nonspecific degradation and acts as a template for synthesis of new cRNA-RNase molecules (step 5). Once the cycle is initiated, there may be no further requirement for nuclear-encoded dsRNA. For clarity the model shows cRNA being produced from one strand of the dsRNA duplex, whereas the complex is capable of producing cRNAs from either strand.
Comment in
- Plant gene silencing regularized.
Bruening G. Bruening G. Proc Natl Acad Sci U S A. 1998 Nov 10;95(23):13349-51. doi: 10.1073/pnas.95.23.13349. Proc Natl Acad Sci U S A. 1998. PMID: 9811802 Free PMC article. No abstract available.
Similar articles
- RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants.
Baulcombe DC. Baulcombe DC. Plant Mol Biol. 1996 Oct;32(1-2):79-88. doi: 10.1007/BF00039378. Plant Mol Biol. 1996. PMID: 8980475 Review. - A paragenetic perspective on integration of RNA silencing into the epigenome and its role in the biology of higher plants.
Jorgensen RA, Doetsch N, Müller A, Que Q, Gendler K, Napoli CA. Jorgensen RA, et al. Cold Spring Harb Symp Quant Biol. 2006;71:481-5. doi: 10.1101/sqb.2006.71.023. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381330 Review. - Cosuppression of I transposon activity in Drosophila by I-containing sense and antisense transgenes.
Jensen S, Gassama MP, Heidmann T. Jensen S, et al. Genetics. 1999 Dec;153(4):1767-74. doi: 10.1093/genetics/153.4.1767. Genetics. 1999. PMID: 10581283 Free PMC article. - Methods for engineering resistance to plant viruses.
Sudarshana MR, Roy G, Falk BW. Sudarshana MR, et al. Methods Mol Biol. 2007;354:183-95. doi: 10.1385/1-59259-966-4:183. Methods Mol Biol. 2007. PMID: 17172755 Review. - Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences.
Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Jorgensen RA, et al. Plant Mol Biol. 1996 Aug;31(5):957-73. doi: 10.1007/BF00040715. Plant Mol Biol. 1996. PMID: 8843939
Cited by
- Integrative computational epigenomics to build data-driven gene regulation hypotheses.
Chen T, Tyagi S. Chen T, et al. Gigascience. 2020 Jun 1;9(6):giaa064. doi: 10.1093/gigascience/giaa064. Gigascience. 2020. PMID: 32543653 Free PMC article. - Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes.
Parent JS, Jauvion V, Bouché N, Béclin C, Hachet M, Zytnicki M, Vaucheret H. Parent JS, et al. Nucleic Acids Res. 2015 Sep 30;43(17):8464-75. doi: 10.1093/nar/gkv753. Epub 2015 Jul 24. Nucleic Acids Res. 2015. PMID: 26209135 Free PMC article. - Development of a transgenic early flowering pear (Pyrus communis L.) genotype by RNAi silencing of PcTFL1-1 and PcTFL1-2.
Freiman A, Shlizerman L, Golobovitch S, Yablovitz Z, Korchinsky R, Cohen Y, Samach A, Chevreau E, Le Roux PM, Patocchi A, Flaishman MA. Freiman A, et al. Planta. 2012 Jun;235(6):1239-51. doi: 10.1007/s00425-011-1571-0. Epub 2011 Dec 28. Planta. 2012. PMID: 22203321 - Transgenic resistance to Citrus tristeza virus in grapefruit.
Febres VJ, Lee RF, Moore GA. Febres VJ, et al. Plant Cell Rep. 2008 Jan;27(1):93-104. doi: 10.1007/s00299-007-0445-1. Epub 2007 Sep 20. Plant Cell Rep. 2008. PMID: 17882423
References
- Powell P A, Sanders P R, Tumer N, Fraley R T, Beachy R N. Virology. 1990;175:124–130. - PubMed
- Lindbo J A, Dougherty W G. Mol Plant–Microbe Interact. 1992;5:144–153. - PubMed
- Lindbo J A, Dougherty W G. Virology. 1992;189:725–733. - PubMed
- Stam M, Mol J N M, Kooter J M. Ann Bot. 1997;79:3–12.
LinkOut - more resources
Full Text Sources
Other Literature Sources