Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes - PubMed (original) (raw)
Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes
B Fahrenkrog et al. J Cell Biol. 1998.
Abstract
The nuclear pore complex (NPC), a supramolecular assembly of approximately 100 different proteins (nucleoporins), mediates bidirectional transport of molecules between the cytoplasm and the cell nucleus. Extensive structural studies have revealed the three- dimensional (3D) architecture of Xenopus NPCs, and eight of the approximately 12 cloned and characterized vertebrate nucleoporins have been localized within the NPC. Thanks to the power of yeast genetics, 30 yeast nucleoporins have recently been cloned and characterized at the molecular level. However, the localization of these nucleoporins within the 3D structure of the NPC has remain elusive, mainly due to limitations of preparing yeast cells for electron microscopy (EM). We have developed a new protocol for preparing yeast cells for EM that yielded structurally well-preserved yeast NPCs. A direct comparison of yeast and Xenopus NPCs revealed that the NPC structure is evolutionarily conserved, although yeast NPCs are 15% smaller in their linear dimensions. With this preparation protocol and yeast strains expressing nucleoporins tagged with protein A, we have localized Nsp1p and its interacting partners Nup49p, Nup57p, Nup82p, and Nic96p by immuno-EM. Accordingly, Nsp1p resides in three distinct subcomplexes which are located at the entry and exit of the central gated channel and at the terminal ring of the nuclear basket.
Figures
Figure 2
Schematic diagram of the fusion constructs used in this study: (A) Four IgG-binding domains derived from S. aureus protein A (ProtA) and the essential NSP1 COOH-terminal domain. (B) Two IgG-binding domains of ProtA and the NIC96 gene under control of the NOP1 promoter and URA3 selection. (C) Four IgG-binding domains of ProtA and the NUP49 gene. (D) Two IgG-binding domains of ProtA and the NUP57 gene under control of the NOP1 promotor and HIS3 selection. (E) Two IgG-binding domains of ProtA and the NUP82 gene under control of the NOP1 promotor and HIS3 selection. (F) Two IgG-binding domains of ProtA and the PUS1 gene under control of the NOP1 promotor and HIS3 selection. (g) Two IgG-binding domains of ProtA and the DHFR gene under control of the NOP1 promotor and URA3 selection. Right, abbreviations for the yeast strains carrying protein A–tagged nucleoporins used in this paper.
Figure 1
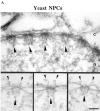
Yeast NPCs. (A) Cross section through the NE and a gallery of selected electron micrographs of NPC cross sections. Large arrowheads, nuclear baskets; small arrowheads, cytoplasmic fibrils; c, cytoplasm; n, nucleus. (B) Schematic comparison of Xenopus and yeast NPCs. Bar, 100 nm.
Figure 1
Yeast NPCs. (A) Cross section through the NE and a gallery of selected electron micrographs of NPC cross sections. Large arrowheads, nuclear baskets; small arrowheads, cytoplasmic fibrils; c, cytoplasm; n, nucleus. (B) Schematic comparison of Xenopus and yeast NPCs. Bar, 100 nm.
Figure 3
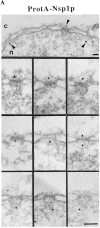
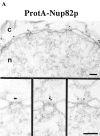
Immunogold localization of Nsp1p in ProtA–Nsp1p cells. (A) Triton X-100– extracted spheroplasts from ProtA–Nsp1p cells were pre-immunolabeled with a polyclonal anti–protein A antibody conjugated to 8-nm colloidal gold, and prepared for EM by Epon embedding and thin sectioning. Shown is a view along a cross sectioned NE stretch with labeled NPCs (arrowheads), together with a gallery of selected examples of gold-labeled NPC cross sections. The antibody labeled the cytoplasmic periphery and the nuclear periphery of the central gated channel, and the terminal ring of the nuclear baskets. c, cytoplasm; n, nucleus. (B) Quantitative analysis of the gold particles associated with the NPCs in the ProtA–Nsp1p strain after labeling with gold-conjugated anti–protein A antibody. 88 gold particles were scored. Bars, 100 nm.
Figure 3
Immunogold localization of Nsp1p in ProtA–Nsp1p cells. (A) Triton X-100– extracted spheroplasts from ProtA–Nsp1p cells were pre-immunolabeled with a polyclonal anti–protein A antibody conjugated to 8-nm colloidal gold, and prepared for EM by Epon embedding and thin sectioning. Shown is a view along a cross sectioned NE stretch with labeled NPCs (arrowheads), together with a gallery of selected examples of gold-labeled NPC cross sections. The antibody labeled the cytoplasmic periphery and the nuclear periphery of the central gated channel, and the terminal ring of the nuclear baskets. c, cytoplasm; n, nucleus. (B) Quantitative analysis of the gold particles associated with the NPCs in the ProtA–Nsp1p strain after labeling with gold-conjugated anti–protein A antibody. 88 gold particles were scored. Bars, 100 nm.
Figure 4
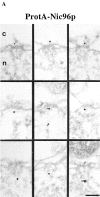
Localization of Nic96p in the ProtA–Nic96p strain. (A) Gallery of selected NPC cross sections showing labeling with the anti–protein A antibody conjugated to 8-nm colloidal gold in Triton X-100–extracted spheroplasts form ProtA–Nic96p cells. Similar to Nsp1p (see Fig. 3), Nic96p is localized at the cytoplasmic (top) and the nuclear periphery (middle) of the central gated channel, and at the terminal ring of the nuclear baskets (bottom). c, cytoplasm; n, nucleus. (B) Quantitation of the gold particle distribution associated with NPCs in the ProtA–Nic96p strain. 80 gold particles were scored. Bar, 100 nm.
Figure 4
Localization of Nic96p in the ProtA–Nic96p strain. (A) Gallery of selected NPC cross sections showing labeling with the anti–protein A antibody conjugated to 8-nm colloidal gold in Triton X-100–extracted spheroplasts form ProtA–Nic96p cells. Similar to Nsp1p (see Fig. 3), Nic96p is localized at the cytoplasmic (top) and the nuclear periphery (middle) of the central gated channel, and at the terminal ring of the nuclear baskets (bottom). c, cytoplasm; n, nucleus. (B) Quantitation of the gold particle distribution associated with NPCs in the ProtA–Nic96p strain. 80 gold particles were scored. Bar, 100 nm.
Figure 5
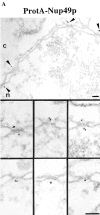
Nup49p (A) and Nup57p (C) are located at the cytoplasmic and the nuclear periphery of the central gated channel. Triton X-100–extracted spheroplasts from the ProtA–Nup49p and Nup57p–ProtA strains were incubated with an anti– protein A antibody conjugated to 8-nm colloidal gold (B and D). c, cytoplasm; n, nucleus. Distribution of the gold particles associated with NPCs in the ProtA–Nup49p and Nup57p–ProtA strains, respectively. 79 and 93 gold particles were scored, respectively. Bars, 100 nm.
Figure 5
Nup49p (A) and Nup57p (C) are located at the cytoplasmic and the nuclear periphery of the central gated channel. Triton X-100–extracted spheroplasts from the ProtA–Nup49p and Nup57p–ProtA strains were incubated with an anti– protein A antibody conjugated to 8-nm colloidal gold (B and D). c, cytoplasm; n, nucleus. Distribution of the gold particles associated with NPCs in the ProtA–Nup49p and Nup57p–ProtA strains, respectively. 79 and 93 gold particles were scored, respectively. Bars, 100 nm.
Figure 5
Nup49p (A) and Nup57p (C) are located at the cytoplasmic and the nuclear periphery of the central gated channel. Triton X-100–extracted spheroplasts from the ProtA–Nup49p and Nup57p–ProtA strains were incubated with an anti– protein A antibody conjugated to 8-nm colloidal gold (B and D). c, cytoplasm; n, nucleus. Distribution of the gold particles associated with NPCs in the ProtA–Nup49p and Nup57p–ProtA strains, respectively. 79 and 93 gold particles were scored, respectively. Bars, 100 nm.
Figure 5
Nup49p (A) and Nup57p (C) are located at the cytoplasmic and the nuclear periphery of the central gated channel. Triton X-100–extracted spheroplasts from the ProtA–Nup49p and Nup57p–ProtA strains were incubated with an anti– protein A antibody conjugated to 8-nm colloidal gold (B and D). c, cytoplasm; n, nucleus. Distribution of the gold particles associated with NPCs in the ProtA–Nup49p and Nup57p–ProtA strains, respectively. 79 and 93 gold particles were scored, respectively. Bars, 100 nm.
Figure 6
Nup82p is found only at the cytoplasmic periphery of the central gated channel. (A) Triton X-100–extracted spheroplasts from ProtA–Nup82p cells were pre-immunolabeled with an anti–protein A antibody directly conjugated to 8-nm gold. Shown are a stretch along the NE and a gallery of selected examples of gold-labeled NPC cross sections. c, cytoplasm; n, nucleus. Bars, 100 nm. (B) The quantitative analysis of the gold particle distribution revealed that Nup82p resides in the center of the NPC in a distance of ∼40 nm from the central plane of the NPC. 106 gold particles were scored for the quantitative analysis.
Figure 6
Nup82p is found only at the cytoplasmic periphery of the central gated channel. (A) Triton X-100–extracted spheroplasts from ProtA–Nup82p cells were pre-immunolabeled with an anti–protein A antibody directly conjugated to 8-nm gold. Shown are a stretch along the NE and a gallery of selected examples of gold-labeled NPC cross sections. c, cytoplasm; n, nucleus. Bars, 100 nm. (B) The quantitative analysis of the gold particle distribution revealed that Nup82p resides in the center of the NPC in a distance of ∼40 nm from the central plane of the NPC. 106 gold particles were scored for the quantitative analysis.
Figure 7
In vivo location of GFP-tagged Nsp1p in nup49-313 cells. NUP49+ and nup49-313 cells transformed with a single copy ARS/CEN/ADE2 plasmid harboring a fusion construct between GFP and the Nsp1p COOH-terminal domain were grown at permissive temperature (20_°_C) or shifted for 6 h to restrictive temperature (37_°_C). In the case of the nup49-313 strain, cells were regrown after a 6-h shift to 37°C for a further 2 h at the permissive temperature (20_°_C). Cells were inspected in the fluorescence microscope to detect the GFP fusion protein and also viewed by Nomarski optics.
Figure 8
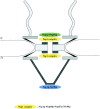
Schematic representation of the yeast NPC with the 3D localization of three Nsp1p subcomplexes. The Nsp1p–Nup49p–Nup57p– Nic96p complex (i.e., the Nsp1p complex) is localized at both the cytoplasmic and nuclear periphery of the central gated channel, and the Nsp1p–Nup82p complex resides only at the cytoplasmic periphery of the central gated channel. In addition, a third Nsp1p complex consisting of Nsp1p interacting with Nic96p is located at the terminal ring of the nuclear baskets. c, cytoplasmic, and n, nuclear side of the NE.
Similar articles
- Comparative spatial localization of protein-A-tagged and authentic yeast nuclear pore complex proteins by immunogold electron microscopy.
Fahrenkrog B, Aris JP, Hurt EC, Panté N, Aebi U. Fahrenkrog B, et al. J Struct Biol. 2000 Apr;129(2-3):295-305. doi: 10.1006/jsbi.2000.4223. J Struct Biol. 2000. PMID: 10806080 - The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport.
Bailer SM, Balduf C, Hurt E. Bailer SM, et al. Mol Cell Biol. 2001 Dec;21(23):7944-55. doi: 10.1128/MCB.21.23.7944-7955.2001. Mol Cell Biol. 2001. PMID: 11689687 Free PMC article. - A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p.
Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. Grandi P, et al. J Cell Biol. 1995 Sep;130(6):1263-73. doi: 10.1083/jcb.130.6.1263. J Cell Biol. 1995. PMID: 7559750 Free PMC article. - [Nuclear pores: from yeast to higher eukaryotes].
Doye V. Doye V. J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French. - The nuclear pore complex: from molecular architecture to functional dynamics.
Stoffler D, Fahrenkrog B, Aebi U. Stoffler D, et al. Curr Opin Cell Biol. 1999 Jun;11(3):391-401. doi: 10.1016/S0955-0674(99)80055-6. Curr Opin Cell Biol. 1999. PMID: 10395558 Review.
Cited by
- Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay.
Miller BR, Powers M, Park M, Fischer W, Forbes DJ. Miller BR, et al. Mol Biol Cell. 2000 Oct;11(10):3381-96. doi: 10.1091/mbc.11.10.3381. Mol Biol Cell. 2000. PMID: 11029043 Free PMC article. - Importin beta-depending nuclear import pathways: role of the adapter proteins in the docking and releasing steps.
Rollenhagen C, Mühlhäusser P, Kutay U, Panté N. Rollenhagen C, et al. Mol Biol Cell. 2003 May;14(5):2104-15. doi: 10.1091/mbc.e02-06-0372. Epub 2003 Feb 6. Mol Biol Cell. 2003. PMID: 12802078 Free PMC article. - In situ analysis of spatial relationships between proteins of the nuclear pore complex.
Damelin M, Silver PA. Damelin M, et al. Biophys J. 2002 Dec;83(6):3626-36. doi: 10.1016/S0006-3495(02)75363-0. Biophys J. 2002. PMID: 12496130 Free PMC article. - Functional characterization of a Nup159p-containing nuclear pore subcomplex.
Belgareh N, Snay-Hodge C, Pasteau F, Dagher S, Cole CN, Doye V. Belgareh N, et al. Mol Biol Cell. 1998 Dec;9(12):3475-92. doi: 10.1091/mbc.9.12.3475. Mol Biol Cell. 1998. PMID: 9843582 Free PMC article. - Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction.
Romes EM, Tripathy A, Slep KC. Romes EM, et al. J Biol Chem. 2012 May 4;287(19):15862-73. doi: 10.1074/jbc.M111.336172. Epub 2012 Mar 12. J Biol Chem. 2012. PMID: 22411995 Free PMC article.
References
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. - PubMed
- Barth W, Stochaj U. The yeast nucleoporin Nsp1 binds nuclear localization sequences in vitro. . Biochem Cell Biol. 1996;74:363–372. - PubMed
- Baschong W, Wrigley NG. Small colloidal gold conjugated to Fab fragments or to immunoglobulin G as high-resolution labels for electron microscopy: a technical overview. J Electron Microsc Technol. 1990;14:313–323. - PubMed
- Carmo-Fonseca M, Kern H, Hurt EC. Human nucleoporin p62 and the essential yeast nuclear pore protein NSP1 show sequence homology and a similar domain organization. Eur J Cell Biol. 1991;55:17–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases