Light chain-dependent regulation of Kinesin's interaction with microtubules - PubMed (original) (raw)
Light chain-dependent regulation of Kinesin's interaction with microtubules
K J Verhey et al. J Cell Biol. 1998.
Abstract
We have investigated the mechanism by which conventional kinesin is prevented from binding to microtubules (MTs) when not transporting cargo. Kinesin heavy chain (HC) was expressed in COS cells either alone or with kinesin light chain (LC). Immunofluorescence microscopy and MT cosedimentation experiments demonstrate that the binding of HC to MTs is inhibited by coexpression of LC. Association between the chains involves the LC NH2-terminal domain, including the heptad repeats, and requires a region of HC that includes the conserved region of the stalk domain and the NH2 terminus of the tail domain. Inhibition of MT binding requires in addition the COOH-terminal 64 amino acids of HC. Interaction between the tail and the motor domains of HC is supported by sedimentation experiments that indicate that kinesin is in a folded conformation. A pH shift from 7.2 to 6.8 releases inhibition of kinesin without changing its sedimentation behavior. Endogenous kinesin in COS cells also shows pH-sensitive inhibition of MT binding. Taken together, our results provide evidence that a function of LC is to keep kinesin in an inactive ground state by inducing an interaction between the tail and motor domains of HC; activation for cargo transport may be triggered by a small conformational change that releases the inhibition of the motor domain for MT binding.
Figures
Figure 1
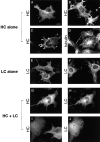
Localization of HC expressed alone, LC expressed alone, and HC and LC expressed together. COS cells were transiently transfected with plasmids encoding HC alone (A–D), LC alone (E and F) or both HC and LC (G–J). The expressed proteins were localized by immunofluorescence microscopy. (A and B) HC was detected with an anti–myc monoclonal antibody followed by Rhodamine Red-X–labeled anti–mouse secondary antibody. (C and D) Cells were double labeled for HC and MTs. HC was detected with anti–myc polyclonal and Oregon green 488–labeled anti–rabbit antibodies, and tubulin was detected with antitubulin monoclonal and Rhodamine Red-X–labeled anti–mouse secondary antibodies. (E and F) LC was detected with anti–HA polyclonal and Oregon green 488–labeled anti–rabbit secondary antibodies. (G–J) Cells were double labeled for HC and LC. HC was detected with anti–myc monoclonal and Rhodamine Red-X–labeled anti–mouse antibodies, and LC was detected with anti–HA polyclonal and Oregon green 488– labeled anti–rabbit secondary antibodies. Bar, 10 μm.
Figure 3
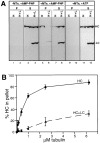
Interaction of the LC deletion mutants with HC. (A) Deletion mutants of LC were constructed in which stop codons were engineered after amino acids 176, 237, 405, and 488. These constructs retain the heptad repeat region but contain deletions of some or all of the TPR motifs. The deletion mutant LΔHR is missing only the heptad repeat region. Amino acids numbers for the full length protein are shown below the schematic of LC. (B) COS cells expressing HC, LC, or the LC deletion mutants alone (H, L, L176, L237, L405, L488, LΔHR; top) or expressing HC together with LC or the LC deletion mutants (H+L, H+L176, H+L237, H+L405, H+L488, and H+LΔHR; bottom) were lysed with 1% Triton X-100. After centrifugation, the amount of expressed protein found insoluble in the pellet (P) and soluble in the lysate (S) was determined by immunoblotting with polyclonal antibodies to the myc- and HA-epitope tags. Molecular weight size standards (kilodaltons) are indicated on the left. (C) Coimmunoprecipitation of HC and the LC deletion mutants. Lysates were immunoprecipitated for HC using a monoclonal anti–myc antibody (left) or for LC using a monoclonal anti–HA antibody (right). Immunoprecipitates were immunoblotted to detect the expressed proteins using polyclonal antibodies to the epitope tags.
Figure 2
MT cosedimentation of HC expressed alone or together with LC. (A) Lysates from COS cells expressing HC alone (H) or together with LC (H+L) were compared under three conditions: no addition of MTs (left) or addition of taxol-stabilized MTs with either AMP-PNP (middle) or ATP (right). MTs and bound proteins were sedimented through a sucrose cushion and the MT pellets (P) and supernatants (S) were immunoblotted for the presence of the expressed HC and LC. (B) Quantitation of the amount of HC recovered in the MT pellet in the presence of AMP-PNP as a function of the amount of MTs added to the lysate. (•) Data from cells expressing HC alone, (▴) cells coexpressing HC and LC. Each data point represents the mean ± SD of at least four experiments.
Figure 4
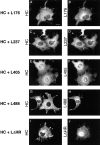
Localization of HC expressed with the LC deletion mutants. COS cells were transiently transfected with plasmids encoding the indicated proteins and the expressed proteins were localized by immunofluorescence microscopy. HC was detected with anti–myc monoclonal and Rhodamine Red-X–labeled anti–mouse secondary antibodies. The LC deletion mutants were detected with anti–HA polyclonal and Oregon green 488–labeled anti–rabbit secondary antibodies. For coexpression of HC with L488, note that the cell on the left expresses both proteins and shows diffuse staining, whereas the cell on the right expresses only HC and shows MT staining (G and H). Bar, 10 μm.
Figure 5
MT cosedimentation of HC expressed with the LC deletion mutants. Taxol-stabilized MTs and AMP-PNP were added to lysates from COS cells expressing HC alone (H), HC and LC (H+L), or HC and the LC deletion mutants (H+L176, H+L237, H+L405, and H+L488). MTs and bound proteins were sedimented through a sucrose cushion. The MT pellets (P) and supernatants (S) were immunoblotted to detect the expressed proteins using polyclonal antibodies to the myc- and HA-tags.
Figure 6
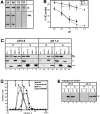
Interaction of the HC deletion mutants with LC. (A) Deletion mutants of HC were constructed in which stop codons were engineered after amino acids 682, 810, and 891. These deletions remove both the tail domain and the conserved region of the stalk domain (H682), the entire tail domain (H810), or half of the tail domain (H891). (B) COS cells expressing HC or the HC deletion mutants alone (H, H682, H810, and H891; top) or together with LC (H+L, H682+L, H810+L, and H891+L; bottom) were lysed with 1% Triton X-100. After centrifugation, the amount of expressed protein found insoluble in the pellet (P) and soluble in the lysate (S) was determined by immunoblotting with polyclonal antibodies to the myc- and HA-epitope tags. Molecular weight size standards (kilodaltons) are indicated on the left. (C) Coimmunoprecipitation of the HC deletion mutants and LC. Lysates were immunoprecipitated for HC using a monoclonal anti–myc antibody (left) or for LC using a monoclonal anti–HA antibody (right). Immunoprecipitates were immunoblotted to detect the expressed proteins using polyclonal antibodies to the epitope tags.
Figure 7
MT cosedimentation of the HC deletion mutants expressed alone or together with LC. Taxol-stabilized MTs and AMP-PNP were added to lysates from COS cells expressing HC (H) or the deletion mutants (H682, H810, and H891) alone or together with LC (H+L, H682+L, H810+L, and H891+L). MTs and bound proteins were sedimented through a sucrose cushion. The MT pellets (P) and supernatants (S) were immunoblotted to detect the expressed proteins using polyclonal antibodies to the myc- and HA-tags.
Figure 8
MT cosedimentation of expressed recombinant HC+LC and endogenous kinesin is pH dependent. (A) Taxol-stabilized MTs and AMP-PNP were added to lysates from COS cells expressing HC (H) or HC and LC (H+L). Lysates were prepared in LB+Triton X-100 buffered with MES (pH 6.4), Pipes (pH 6.8), or Hepes (pH 7.2 and 7.6). MTs and bound proteins were sedimented through a sucrose cushion and the MT pellets and supernatants were immunoblotted with antibodies against myc- and HA-epitope tags to detect the expressed recombinant proteins. Only the immunoblots of the MT pellets are shown. (B) The pH dependence of MT binding of HC alone (circles) or HC and LC (triangles) was determined between pH 6.8 and 7.2 in LB + Triton X-100 buffered with Pipes (solid line) or Hepes (dashed line). Each data point represents the mean ± SD of at least five separate experiments. (C) Taxol-stabilized MTs and AMP-PNP were added to lysates, prepared at pH 6.8 or 7.2 as in A, from COS cells coexpressing the indicated recombinant proteins. MTs and bound proteins were sedimented through a sucrose cushion. The MT pellets (P) and supernatants (S) were immunoblotted to detect the expressed proteins using polyclonal antibodies to the myc- and HA-tags. (D) Sedimentation analysis of coexpressed HC and LC. COS cell lysates made at pH 6.8 (circles) and 7.2 (squares) were subjected to sedimentation on linear 9–15% sucrose gradients, both at physiological (closed symbols) and high salt (0.5 M NaCl; open symbols) concentrations. The expressed recombinant proteins were detected in the fractions by immunoblotting with polyclonal antibodies to the myc- and HA-tags. The mobility on the gradient of proteins with known S values is indicated. (E) Taxol-stabilized MTs and AMP-PNP were added to lysates, prepared at pH 6.8 or 7.2 as in A, from untransfected COS cells. MTs and bound proteins were sedimented through a sucrose cushion. The MT pellets (P) and supernatants (S) were immunoblotted to detect endogenous kinesin using a polyclonal antibody to kinesin HC.
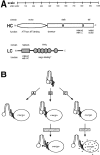
Figure 9
Functional domains of kinesin and models for the activation of kinesin in vivo. (A) Representation of the domains of kinesin HC and LC, drawn to scale, and the functions assigned to them. (B) Possible mechanisms to explain the activation of kinesin for MT binding in vivo. Only one HC and one LC of the kinesin tetramer are shown for simplicity. Soluble kinesin in the cell is in an inactive folded conformation such that the HC tail domain inhibits MT binding of the motor domain. Activation of kinesin, represented here by a hypothetical opening of the folded molecule, may be triggered: (A) by interaction of the motor with its cargo, (B) before or after (as drawn) cargo binding by posttranslational modification (e.g., phosphorylation) of either the HC or LC, or (C) after cargo binding by a localized shift of pH around the vesicle from neutral to slightly acidic.
Similar articles
- Kinesin's light chains inhibit the head- and microtubule-binding activity of its tail.
Wong YL, Rice SE. Wong YL, et al. Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):11781-6. doi: 10.1073/pnas.1005854107. Epub 2010 Jun 14. Proc Natl Acad Sci U S A. 2010. PMID: 20547877 Free PMC article. - The distance that kinesin-1 holds its cargo from the microtubule surface measured by fluorescence interference contrast microscopy.
Kerssemakers J, Howard J, Hess H, Diez S. Kerssemakers J, et al. Proc Natl Acad Sci U S A. 2006 Oct 24;103(43):15812-7. doi: 10.1073/pnas.0510400103. Epub 2006 Oct 11. Proc Natl Acad Sci U S A. 2006. PMID: 17035506 Free PMC article. - Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells.
Navone F, Niclas J, Hom-Booher N, Sparks L, Bernstein HD, McCaffrey G, Vale RD. Navone F, et al. J Cell Biol. 1992 Jun;117(6):1263-75. doi: 10.1083/jcb.117.6.1263. J Cell Biol. 1992. PMID: 1607388 Free PMC article. - These motors were made for walking.
Hunter B, Allingham JS. Hunter B, et al. Protein Sci. 2020 Aug;29(8):1707-1723. doi: 10.1002/pro.3895. Epub 2020 Jun 26. Protein Sci. 2020. PMID: 32472639 Free PMC article. Review. - Review: regulation mechanisms of Kinesin-1.
Adio S, Reth J, Bathe F, Woehlke G. Adio S, et al. J Muscle Res Cell Motil. 2006;27(2):153-60. doi: 10.1007/s10974-005-9054-1. Epub 2006 Feb 1. J Muscle Res Cell Motil. 2006. PMID: 16450053 Review.
Cited by
- Single molecule FRET observation of kinesin-1's head-tail interaction on microtubule.
Aoki T, Tomishige M, Ariga T. Aoki T, et al. Biophysics (Nagoya-shi). 2013 Nov 7;9:149-59. doi: 10.2142/biophysics.9.149. eCollection 2013. Biophysics (Nagoya-shi). 2013. PMID: 27493553 Free PMC article. - Traffic control: regulation of kinesin motors.
Verhey KJ, Hammond JW. Verhey KJ, et al. Nat Rev Mol Cell Biol. 2009 Nov;10(11):765-77. doi: 10.1038/nrm2782. Nat Rev Mol Cell Biol. 2009. PMID: 19851335 Review. - Kinesin molecular motors: transport pathways, receptors, and human disease.
Goldstein LS. Goldstein LS. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):6999-7003. doi: 10.1073/pnas.111145298. Proc Natl Acad Sci U S A. 2001. PMID: 11416178 Free PMC article. Review. - Integrated regulation of motor-driven organelle transport by scaffolding proteins.
Fu MM, Holzbaur EL. Fu MM, et al. Trends Cell Biol. 2014 Oct;24(10):564-74. doi: 10.1016/j.tcb.2014.05.002. Epub 2014 Jun 18. Trends Cell Biol. 2014. PMID: 24953741 Free PMC article. Review. - Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent.
Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Glater EE, et al. J Cell Biol. 2006 May 22;173(4):545-57. doi: 10.1083/jcb.200601067. J Cell Biol. 2006. PMID: 16717129 Free PMC article.
References
- Block SM. Kinesin: what gives? . Cell. 1998a;93:5–8. - PubMed
- Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. - PubMed
- Bloom GS, Endow SA. Motor proteins 1: kinesins. Protein Profile. 1994;1:1059–1116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources