Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli - PubMed (original) (raw)
Comparative Study
Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli
Y W Tang et al. J Clin Microbiol. 1998 Dec.
Abstract
Rapid and accurate identification of bacterial pathogens is a fundamental goal of clinical microbiology, but one that is difficult or impossible for many slow-growing and fastidious organisms. We used identification systems based on cellular fatty acid profiles (Sherlock; MIDI, Inc., Newark, Del.), carbon source utilization (Microlog; Biolog, Inc., Hayward, Calif.), and 16S rRNA gene sequence (MicroSeq; Perkin-Elmer Applied Biosystems Division, Foster City, Calif.) to evaluate 72 unusual aerobic gram-negative bacilli isolated from clinical specimens at the Mayo Clinic. Compared to lengthy conventional methods, Sherlock, Microlog, and MicroSeq were able to identify 56 of 72 (77.8%), 63 of 72 (87.5%), and 70 of 72 (97.2%) isolates to the genus level (P = 0.002) and 44 to 65 (67.7%), 55 of 65 (84.6%), and 58 of 65 (89.2%) isolates to the species level (P = 0.005), respectively. Four Acinetobacter and three Bordetella isolates which could not be identified to the species level by conventional methods were identified by MicroSeq. In comparison to the full 16S rDNA sequences, the first 527 bp provided identical genus information for all 72 isolates and identical species information for 67 (93.1%) isolates. These data show that MicroSeq provides rapid, unambiguous identification of clinical bacterial isolates. The improved turnaround time provided by genotypic identification systems may translate into improved clinical outcomes.
Figures
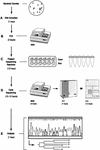
FIG. 1
Flowchart of the MicroSeq process from culture to sequence. The total elapsed time was 15.5 to 18.5 h, comprising bacterial DNA extraction (A), PCR (B), sequencing reaction preparation (C), cycle sequencing (D), and analysis (E). The time required for each step is indicated.
FIG. 2
Neighbor-joining analysis of DNA sequences from several Enterobacter spp. Phylogenetic analysis was based on full 16S rRNA gene sequences, and the scale reflects relative phylogenetic distance. Isolates with names beginning with Mayo were evaluated in this study. Isolates with names beginning with accession numbers were retrieved from GenBank (15, 16). The remaining isolates, whose names begin with ATCC numbers, were type strains stored in the MicroSeq database.
Similar articles
- Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis.
Tang YW, Von Graevenitz A, Waddington MG, Hopkins MK, Smith DH, Li H, Kolbert CP, Montgomery SO, Persing DH. Tang YW, et al. J Clin Microbiol. 2000 Apr;38(4):1676-8. doi: 10.1128/JCM.38.4.1676-1678.2000. J Clin Microbiol. 2000. PMID: 10747168 Free PMC article. - Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically significant bacterial isolates with ambiguous biochemical profiles.
Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, Tam DM, Que TL, Yuen KY. Woo PC, et al. J Clin Microbiol. 2003 May;41(5):1996-2001. doi: 10.1128/JCM.41.5.1996-2001.2003. J Clin Microbiol. 2003. PMID: 12734240 Free PMC article. - Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system.
Patel JB, Leonard DG, Pan X, Musser JM, Berman RE, Nachamkin I. Patel JB, et al. J Clin Microbiol. 2000 Jan;38(1):246-51. doi: 10.1128/JCM.38.1.246-251.2000. J Clin Microbiol. 2000. PMID: 10618095 Free PMC article. - Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.
Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Woo PC, et al. Clin Microbiol Infect. 2008 Oct;14(10):908-34. doi: 10.1111/j.1469-0691.2008.02070.x. Clin Microbiol Infect. 2008. PMID: 18828852 Review. - Contemporary microbiology and identification of Corynebacteria spp. causing infections in human.
Zasada AA, Mosiej E. Zasada AA, et al. Lett Appl Microbiol. 2018 Jun;66(6):472-483. doi: 10.1111/lam.12883. Epub 2018 Apr 17. Lett Appl Microbiol. 2018. PMID: 29573441 Review.
Cited by
- In silico analysis of 16S ribosomal RNA gene sequencing-based methods for identification of medically important anaerobic bacteria.
Woo PC, Chung LM, Teng JL, Tse H, Pang SS, Lau VY, Wong VW, Kam KL, Lau SK, Yuen KY. Woo PC, et al. J Clin Pathol. 2007 May;60(5):576-9. doi: 10.1136/jcp.2006.038653. Epub 2006 Oct 17. J Clin Pathol. 2007. PMID: 17046845 Free PMC article. - Particular Distribution of Enterobacter cloacae Strains Isolated from Urinary Tract Infection within Clonal Complexes.
Akbari M, Bakhshi B, Najar Peerayeh S. Akbari M, et al. Iran Biomed J. 2016;20(1):49-55. doi: 10.7508/ibj.2016.01.007. Epub 2015 Oct 25. Iran Biomed J. 2016. PMID: 26498349 Free PMC article. - 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory.
Bosshard PP, Zbinden R, Abels S, Böddinghaus B, Altwegg M, Böttger EC. Bosshard PP, et al. J Clin Microbiol. 2006 Apr;44(4):1359-66. doi: 10.1128/JCM.44.4.1359-1366.2006. J Clin Microbiol. 2006. PMID: 16597863 Free PMC article. - Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants.
Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG, Vidaver AK. Zinniel DK, et al. Appl Environ Microbiol. 2002 May;68(5):2198-208. doi: 10.1128/AEM.68.5.2198-2208.2002. Appl Environ Microbiol. 2002. PMID: 11976089 Free PMC article. - Isolation and characterization of bacteriocinogenic lactic bacteria from M-Tuba and Tepache, two traditional fermented beverages in México.
de la Fuente-Salcido NM, Castañeda-Ramírez JC, García-Almendárez BE, Bideshi DK, Salcedo-Hernández R, Barboza-Corona JE. de la Fuente-Salcido NM, et al. Food Sci Nutr. 2015 Sep;3(5):434-42. doi: 10.1002/fsn3.236. Epub 2015 Apr 29. Food Sci Nutr. 2015. PMID: 26405529 Free PMC article.
References
- Andersen B M. Biochemical profiles and serotypes of nosocomial Enterobacter cloacae strains in Northern Norway: biochemical identification problems with commercial test systems. Infection. 1995;23:339–343. - PubMed
- Bottger E C, Teske A, Kirschner P, Bost S, Chang H R, Beer V, Hirschel B. Disseminated “Mycobacterium genavense” infection in patients with AIDS. Lancet. 1992;340:76–80. - PubMed
- Bruckner D A, Colonna P. Nomenclature for aerobic and facultative bacteria. Clin Infect Dis. 1997;25:1–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources