Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo - PubMed (original) (raw)
Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo
I Aksan et al. Mol Cell Biol. 1998 Dec.
Abstract
The development of melanocytes, which are pigment-producing cells responsible for skin, hair, and eye color, is absolutely dependent on the action of the microphthalmia basic helix-loop-helix-leucine zipper (bHLH-LZ) transcription factor (Mi); mice lacking a functional Mi protein are entirely devoid of pigment cells. Mi has been shown to activate transcription of the tyrosinase, TRP-1, TRP-2, and QNR-71 genes through specific E-box elements, most notably the highly conserved M box. We investigated the mechanism which enables Mi to be recruited specifically to a restricted subset of E boxes in target promoters while being prevented from binding E-box elements in other promoters. We show both in vitro and in vivo that the presence of a T residue flanking a CATGTG E box is an essential determinant of the ability of Mi to bind DNA, and we successfully predict that the CATGTG E box from the P gene would not bind Mi. In contrast, no specific requirement for the sequences flanking a CACGTG E box was observed, and no binding to an atypical E box in the c-Kit promoter was detected. The relevance of these observations to the control of melanocyte-specific gene expression was highlighted by the fact that the E-box elements located in the tyrosinase, TRP-1, TRP-2, and QNR-71 promoters without exception possess a 5' flanking T residue which is entirely conserved between species as diverse as man and turtle. The ability of Mi to discriminate between different E-box motifs provides a mechanism to restrict the repertoire of genes which are likely to be regulated by Mi and provides insight into the ability of bHLH-LZ transcription factors to achieve the specificity required for the precise coordination of transcription during development.
Figures
FIG. 1
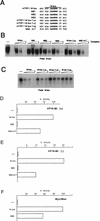
Mi discriminates between highly related E-box motifs. (A) E-box motifs used as probes and competitors. Only the E-box and flanking sequences are shown. The full sequence of each oligonucleotide is given in Table 1. (B) DNA-binding band shift (electrophoretic mobility shift) assay using bacterially expressed and purified Mi together with the M-box probe and the indicated competitor oligonucleotides at 10, 50, and 250 ng. Only the bound DNA is shown. (C) DNA-binding assay using in vitro transcribed and translated Mi and the M-box probe and indicated oligonucleotide competitors at 10, 50, and 250 ng. (D) Yeast one-hybrid assay using a CYC-lacZ reporter containing the indicated target elements, cotransformed with a vector expressing Mi(+) fused to the VP16 activation domain. (E) As for panel D but using a VP16-Mi(−) expression vector. (F) Yeast assay for activity of native Myc and Max proteins on the indicated _CYC-lacZ_-based reporters. Myc and Max were coexpressed in yeast with activation of transcription by Myc being absolutely dependent on Max expression, while Max alone does not activate transcription in this assay (not shown). Residue substitutions are indicated by “greater than” symbols. For example, T>A indicates a T-to-A substitution.
FIG. 2
Conservation of the E-box motifs within melanocyte-specific promoters and between species. The tyrosinase promoter contains three Mi-responsive CATGTG E-box motifs termed the TDE, the M box, and the initiator E box. All three elements from all four species possess a 5′ flanking T residue (boxed) at the −4 position on either the top strand (M box) the bottom strand (initiator E box) or both strands (TDE). The conservation of the 5′ flanking T residue is also seen in the Mi-responsive elements from the quail QNR71 promoter as well as in the TRP-1 M box, and the M box and upstream E box from the TRP-2 promoter. The numbers indicate the positions of the sequences shown relative to the transcription start sites in the cognate promoters.
FIG. 3
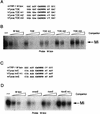
Mi binding to the TDE and initiator (Inr) require the presence of a 5′ flanking T residue. (A, C) E-box motifs used for probes and competitors in the DNA binding band shift assays (for complete sequences see Table 1). (B, D) Band shift assays using an M-box probe and indicated WT and mutant competitors at 10, 50, and 250 ng. Competitors were derived from the mouse TRP-1 M box, the human tyrosinase TDE (hTyros TDE), and the human and mouse initiator elements (hTyros InrE and mTyros InrE, respectively). Only the bound DNA is shown.
FIG. 4
Mi does not bind the E-box element in the P gene. (A) Oligonucleotides used as probes or competitors in the DNA binding band shift assay. Full sequences are given in Table 1. (B) Band shift assay using the M box probe and indicated competitors at 10, 50, and 250 ng. Only bound DNA is shown.
FIG. 5
Efficient binding by USF-1 to a CATGTG E box requires a 5′ flanking T residue. (A) WT and mutant oligonucleotides used as probes and competitors. (B) In vitro transcribed-translated USF-1 was used in DNA binding band shift assay using an M-box probe and the indicated competitors at 10, 50, and 250 ng. Unprogrammed reticulocyte lysate was used as a control and the complex formed using the programmed lysate is supershifted by the addition of anti-USF-1 antibody. Only the bound DNA is shown.
FIG. 6
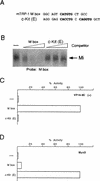
The atypical E-box elements in the c-Kit promoter do not bind Mi. (A) Oligonucleotides used as probe and competitors in the DNA binding assay. (B) Band shift assay using an M-box probe and bacterially expressed and purified Mi together with the indicated competitors at 10, 50, and 250 ng. (C) Yeast one-hybrid assay for binding of VP16 Mi to the M box and c-Kit E-box elements. (D) Yeast one-hybrid assay for binding of MyoD to the M box and c-Kit E-box elements. Yeast were transformed with the indicated CYC-lacZ reporters together with vectors expressing either an Mi-VP16 fusion protein or MyoD were assayed for β-galactosidase activity. The sequences of the oligonucleotides cloned into CYC-lacZ reporter plasmids are listed in Table 1.
Similar articles
- Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene.
Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Yasumoto K, et al. Mol Cell Biol. 1994 Dec;14(12):8058-70. doi: 10.1128/mcb.14.12.8058-8070.1994. Mol Cell Biol. 1994. PMID: 7969144 Free PMC article. - Transcriptional activation of the melanocyte-specific genes by the human homolog of the mouse Microphthalmia protein.
Yasumoto K, Mahalingam H, Suzuki H, Yoshizawa M, Yokoyama K. Yasumoto K, et al. J Biochem. 1995 Nov;118(5):874-81. doi: 10.1093/jb/118.5.874. J Biochem. 1995. PMID: 8749302 - TFE3, a transcription factor homologous to microphthalmia, is a potential transcriptional activator of tyrosinase and TyrpI genes.
Verastegui C, Bertolotto C, Bille K, Abbe P, Ortonne JP, Ballotti R. Verastegui C, et al. Mol Endocrinol. 2000 Mar;14(3):449-56. doi: 10.1210/mend.14.3.0428. Mol Endocrinol. 2000. PMID: 10707962 - Evidence to suggest that expression of MITF induces melanocyte differentiation and haploinsufficiency of MITF causes Waardenburg syndrome type 2A.
Tachibana M. Tachibana M. Pigment Cell Res. 1997 Feb-Apr;10(1-2):25-33. doi: 10.1111/j.1600-0749.1997.tb00462.x. Pigment Cell Res. 1997. PMID: 9170159 Review. - A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work.
Yasumoto K, Amae S, Udono T, Fuse N, Takeda K, Shibahara S. Yasumoto K, et al. Pigment Cell Res. 1998 Dec;11(6):329-36. doi: 10.1111/j.1600-0749.1998.tb00491.x. Pigment Cell Res. 1998. PMID: 9870544 Review.
Cited by
- Melanogenic Inhibition and Toxicity Assessment of Flavokawain A and B on B16/F10 Melanoma Cells and Zebrafish (Danio rerio).
Mohd Sakeh N, Md Razip NN, Mohd Ma'in FI, Abdul Bahari MN, Latif N, Akhtar MN, Balia Yusof ZN, Ahmad S. Mohd Sakeh N, et al. Molecules. 2020 Jul 28;25(15):3403. doi: 10.3390/molecules25153403. Molecules. 2020. PMID: 32731323 Free PMC article. - Hypoxia-inducible factor 1{alpha} is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells.
Buscà R, Berra E, Gaggioli C, Khaled M, Bille K, Marchetti B, Thyss R, Fitsialos G, Larribère L, Bertolotto C, Virolle T, Barbry P, Pouysségur J, Ponzio G, Ballotti R. Buscà R, et al. J Cell Biol. 2005 Jul 4;170(1):49-59. doi: 10.1083/jcb.200501067. Epub 2005 Jun 27. J Cell Biol. 2005. PMID: 15983061 Free PMC article. - MITF controls the TCA cycle to modulate the melanoma hypoxia response.
Louphrasitthiphol P, Ledaki I, Chauhan J, Falletta P, Siddaway R, Buffa FM, Mole DR, Soga T, Goding CR. Louphrasitthiphol P, et al. Pigment Cell Melanoma Res. 2019 Nov;32(6):792-808. doi: 10.1111/pcmr.12802. Epub 2019 Jul 8. Pigment Cell Melanoma Res. 2019. PMID: 31207090 Free PMC article. - The post-translational regulation of transcription factor EB (TFEB) in health and disease.
Takla M, Keshri S, Rubinsztein DC. Takla M, et al. EMBO Rep. 2023 Nov 6;24(11):e57574. doi: 10.15252/embr.202357574. Epub 2023 Sep 20. EMBO Rep. 2023. PMID: 37728021 Free PMC article. Review. - Defective co-activator recruitment in osteoclasts from microphthalmia-oak ridge mutant mice.
Sharma SM, Sif S, Ostrowski MC, Sankar U. Sharma SM, et al. J Cell Physiol. 2009 Jul;220(1):230-7. doi: 10.1002/jcp.21755. J Cell Physiol. 2009. PMID: 19288495 Free PMC article.
References
- Baldwin C T, Lipsky N R, Hoth C F, Cohen T, Mamuya W, Milunsky A. Mutations in PAX3 associated with Waardenburg syndrome type I. Hum Mutat. 1994;3:205–211. - PubMed
- Baynash A G, Hosoda K, Giaid A, Richardson J A, Emoto N, Hammer R E, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases