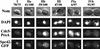Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p - PubMed (original) (raw)
Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase cdc5p
L Cheng et al. Mol Cell Biol. 1998 Dec.
Abstract
Progression through and completion of mitosis require the actions of the evolutionarily conserved Polo kinase. We have determined that the levels of Cdc5p, a Saccharomyces cerevisiae member of the Polo family of mitotic kinases, are cell cycle regulated. Cdc5p accumulates in the nuclei of G2/M-phase cells, and its levels decline dramatically as cells progress through anaphase and begin telophase. We report that Cdc5p levels are sensitive to mutations in key components of the anaphase-promoting complex (APC). We have determined that Cdc5p-associated kinase activity is restricted to G2/M and that this activity is posttranslationally regulated. These results further link the actions of the APC to the completion of mitosis and suggest possible roles for Cdc5p during progression through and completion of mitosis.
Figures
FIG. 1
Decrease in Cdc5p levels as cells exit mitosis. CDC5-ProA cdc 15-2 (YCH236) cells were synchronized in telophase by growth at 37°C for 3 h prior to release into fresh medium at 23°C. Samples for immunoblot analysis of Cdc5-ProA and actin, FACS analysis of DNA content, and detection of anaphase spindles by immunofluorescence staining of tubulin were taken at the times (in minutes) indicated. The percentage of cells with anaphase spindles is shown on the left side of the FACS analysis profile. Spindle morphology in cells was determined by indirect immunofluorescence staining with an antitubulin antibody. Actin detection was used as an internal loading control. Cells released from a cdc15-2 block are delayed in cytokinesis which results in a 2_n and 4_n shift in DNA content after replication.
FIG. 2
The degradation of Cdc5p is dependent on Cdc23p function. Wild-type and cdc23-1 cells were transformed with a 2μm-based plasmid carrying GAL1-HA-CDC5 (PCH740). For both strains, an overnight culture was grown selectively at 23°C in SC lacking histidine. This was inoculated into YEP plus raffinose, grown to early log phase at 23°C and arrested in G1 with α-factor. When >90% of the cells exhibited a schmoo-like morphology, as determined by microscopy, the temperature of the culture was shifted to 37°C (the restrictive temperature for cdc23-1). FACS analysis on these blocked samples also indicated that >90% of the cells had a 1_n_ content of DNA and were in G1 (data not shown). After 30 min at the restrictive temperature, galactose was added for 15 min to induce expression of HA-CDC5 from the GAL promoter. Subsequently, at time 0, glucose was added to turn off expression of HA-CDC5 from the GAL promoter while maintaining the G1 arrest at the restrictive temperature. Samples of equal density were taken at the designated time points and assayed by immunoblotting for HA-Cdc5p and actin. Actin detection was used as an internal loading control.
FIG. 3
Detection of Cdc5p by immunofluorescence in mitotic cells. Cells derived from asynchronous cultures were fixed with methanol and formaldehyde and processed for indirect immunofluorescence. (Left) Population of asynchronous Cdc5-ProA cells (YCH194) stained with anti-IgG to detect Cdc5-ProA. (Right) 4′,6-Diamidino-2-phenylindole (DAPI) to detect DNA using Nomarski optics (NOM-DAPI). Bar = 10 μm.
FIG. 4
Nuclear staining of Cdc5p in asynchronous Cdc5-ProA SPC42-GFP (YCH301) cells. Four views of each cell are shown: Nomarski optics (Nom), staining of DNA (DAPI), Cdc5p-ProA staining (Texas red), and Spc42-GFP using direct fluorescence. On the basis of these four characteristics, cells were placed into the following four groups. I (single G1 cells), II (budded cells with a single nucleus and duplicated SPBs; IIa without and IIb with Cdc5p staining), III (cells in anaphase with nuclei in the mother and daughter neck and separate SPBs present in both mother and daughter), IV (cells which have undergone anaphase, with separate nuclei and polar positioning of SPBs; IVa with and IVb without Cdc5p staining). For each class of cells at given points in the cell cycle, the number and fraction of cells with (IIb, III, and IVa) or without (I, IIa, and IVb) Cdc5p staining are indicated. All images were taken with a 100× objective and printed at the same magnification. Bar = 5 μm.
FIG. 5
Cdc5p is present in cells blocked in S and M phase as detected by immunofluorescence. Wild-type and cdc mutant cells were synchronized at different stages of the cell cycle using either chemicals or by growth of the cdc mutant at the restrictive temperature of 37°C and processed for immunofluorescence. Four views of each cell are shown: Nomarski optics (NOM), staining of DNA (DAPI), Cdc5p-ProA staining (Texas red), and Spc42-GFP (direct fluorescence). The cells were synchronized at the different stages as follows: late G1, with α-factor (YCH301) and cdc4-1 (YCH303); S phase, with HU (YCH301); Metaphase, cdc13-1 (YCH309), nocodazole (Na) (YCH301),and cdc23-1 (YCH305). Samples of the arrested cells used in this study were also used for FACS, immunoblot, and kinase assay analyses shown in Fig. 7. All images were taken with a 100× objective and printed at the same magnification. Bar = 5 μm.
FIG. 6
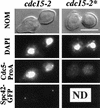
Cdc5p is present in cells synchronized in telophase as detected by immunofluorescence. _CDC5-_ProA cdc15-1 cells (YCH307) were synchronized in telophase by growth at the restrictive temperature and processed for immunofluorescence. CDC5 and SPC42 are not tagged in cdc15-2* cells (YCH238). Four views of each cell are shown: Nomarski optics (NOM), staining of DNA (DAPI), Cdc5p-ProA staining (Texas red), and Spc42-GFP (direct fluorescence). All images were taken with a 100× objective and printed at the same magnification. Bar = 5 μm. ND, not done.
FIG. 7
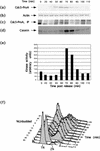
Cdc5p is detected in late G1; S- and M-phase synchronized cells, but Cdc5p-associated kinase activity is restricted to cells arrested in M Phase. Wild-type and cdc mutant cells were arrested in specific stages of the cell cycle using either chemicals or by growth at the restrictive temperature of 37°C, respectively. The cells were synchronized at the various stages as follows: late G1, with α-factor (YCH301) and cdc4-1 (YCH303); G1/S, cells were initially blocked in G1 with α-factor (YCH199) and then released into fresh medium containing HU; S phase, with HU (YCH301); metaphase (Meta), with cdc13-1 (YCH309), nocodazole (NZ) (YCH301), and cdc23-1 (YCH305); and telophase (Tel.), with cdc15-2 (YCH307) and cdc5-1 (YCH214). Extracts from asynchronous (Async) cells were derived from cultures of Cdc5-ProA (YCH199) and strain W303a expressing an untagged Cdc5p. Samples for FACS analysis and extract preparation were taken when >95% of the cells in the culture were appropriately arrested, as detected by light microscopy. Cdc5-ProA (a) and actin (b) in the crude extract were detected by immunoblotting with anti-IgG and antiactin antibodies, respectively. (c) Cdc5-ProA was routinely immunoprecipitated from 400 μg of extract with IgG-Sepharose beads and detected as above by immunoblotting. (d) Kinase activity in these immunocomplexes was measured as described in Materials and Methods with casein as the substrate, except in the case of the 4× HU lane, where 1.6 mg of extract was used. (e) A bar graph of the [32P]-casein levels is shown. Levels were quantitated with a Molecular Dynamics PhosphorImager. (f) Cdc5-ProA kinase activity was determined for cdc5-1-proA (YCH214) cultures grown either asynchronously (Async) (23°C), in the presence of nocodazole (NZ) (3 h, 23°C) or in the presence of nocodazole (3 h, 23°C) followed by a 1-h shift to 37°C (NZ→37). Cdc5-ProA kinase activity was also determined for a wild-type strain expressing Cdc5-ProA (YCH199) arrested in the presence of nocodazole and loaded on the same gel. (g) FACS analysis of DNA content.
FIG. 8
Cdc5p levels and Cdc5p-associated kinase activity fluctuate during the cell cycle. _CDC5-_ProA bar1 (YCH199) cells were synchronized in G1 by addition of α-factor and released into fresh medium lacking α-factor at 23°C. Samples for FACS analysis and extract preparation were taken at the time intervals shown. Cdc5-ProA (a) and (b) actin were detected by immunoblotting with anti-IgG and antiactin antibodies, respectively. (c) Cdc5-ProA was immunoprecipitated (IP) from 400 μg of extract using IgG-Sepharose beads, (d) Kinase activity in the immunocomplexes was measured as described in Materials and Methods with casein as the substrate. (e) A bar graph of the [32P]casein levels is shown. Levels were quantitated with a Molecular Dynamics PhosphorImager. (f) FACS analysis of DNA content.
FIG. 9
Modification of Cdc5p in cdc13-1 cells grown at the nonpermissive temperature is dependent on the DNA damage checkpoint genes, MEC1, RAD53/MEC2, and RAD9. (a) cdc13-1 (YCH216) cells were incubated at 37°C, the nonpermissive temperature for 8 h. Samples for budding index determination and extract preparation were taken at the time intervals shown. (b) cdc13-1 rad53-21 (YCH318), cdc13-1 mec1-1 (YCH317), cdc13-1 rad9::URA3 (YCH316) (rad9-n) and cdc13-1 (YCH216) [**+**]) cells were incubated at 37°C, the nonpermissive temperature for 3 h. cdc13-1 is the only temperature-sensitive mutation in these strains. Samples for extract preparation were taken at 3 h. (c) cdc13-1 rad 53-21 (YCH318), cdc13-1 mec1-1 (YCH317), and cdc13-1 rad9::URA3 (YCH316) (rad9-n) cells were incubated at 37°C, the nonpermissive temperature for 7 h. Samples for extract preparation were taken at 0 and 7 h. Cdc5-ProA and actin were detected by immunoblotting with anti-IgG and antiactin antibodies, respectively.
FIG. 10
Cdc5p-associated kinase activity is phosphorylation dependent. Cdc5-ProA cells (YCH199) were synchronized in metaphase by treatment with nocodazole. Cdc5-ProA was immunoprrecipitated from extract by using IgG-Sepharose, washed into kinase buffer, and divided equally among three samples. (a) CIP (100 U) was either not added (−), added (+), or added to the sample as a control after the CIP was boiled for 10 min to remove phosphatase activity (+*). All three tubes were placed at 37°C for 30 min. The samples were spun down, CIP was washed out using kinase buffer, [γ-32P]ATP and cold ATP were added, and a kinase assay was performed at 30°C. The samples were loaded onto a SDS–9% polyacrylamide gel, and immunoblot analysis was performed using anti-IgG (α IgG) to detect Cdc5-ProA. After color development of the immunoblot (left panel), the filter was exposed to film (right panel). (b) Kinase assays were performed as described above for panel a, except that the phosphatase reactions were carried out in the absence (−) or presence (+) of CIP and in the absence (−) or presence (+) (200 mM) of the CIP inhibitor β-glycerophosphate (β-glyc.).
Similar articles
- Cell cycle regulation of DNA replication initiator factor Dbf4p.
Cheng L, Collyer T, Hardy CF. Cheng L, et al. Mol Cell Biol. 1999 Jun;19(6):4270-8. doi: 10.1128/MCB.19.6.4270. Mol Cell Biol. 1999. PMID: 10330168 Free PMC article. - The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae.
Shirayama M, Zachariae W, Ciosk R, Nasmyth K. Shirayama M, et al. EMBO J. 1998 Mar 2;17(5):1336-49. doi: 10.1093/emboj/17.5.1336. EMBO J. 1998. PMID: 9482731 Free PMC article. - Chromosome separation and exit from mitosis in budding yeast: dependence on growth revealed by cAMP-mediated inhibition.
Anghileri P, Branduardi P, Sternieri F, Monti P, Visintin R, Bevilacqua A, Alberghina L, Martegani E, Baroni MD. Anghileri P, et al. Exp Cell Res. 1999 Aug 1;250(2):510-23. doi: 10.1006/excr.1999.4531. Exp Cell Res. 1999. PMID: 10413604 - The family of polo-like kinases.
Golsteyn RM, Lane HA, Mundt KE, Arnaud L, Nigg EA. Golsteyn RM, et al. Prog Cell Cycle Res. 1996;2:107-14. doi: 10.1007/978-1-4615-5873-6_11. Prog Cell Cycle Res. 1996. PMID: 9552388 Review. - The anaphase-promoting complex: it's not just for mitosis any more.
Harper JW, Burton JL, Solomon MJ. Harper JW, et al. Genes Dev. 2002 Sep 1;16(17):2179-206. doi: 10.1101/gad.1013102. Genes Dev. 2002. PMID: 12208841 Review. No abstract available.
Cited by
- Cell cycle regulation of DNA replication initiator factor Dbf4p.
Cheng L, Collyer T, Hardy CF. Cheng L, et al. Mol Cell Biol. 1999 Jun;19(6):4270-8. doi: 10.1128/MCB.19.6.4270. Mol Cell Biol. 1999. PMID: 10330168 Free PMC article. - Dbf4 regulates the Cdc5 Polo-like kinase through a distinct non-canonical binding interaction.
Chen YC, Weinreich M. Chen YC, et al. J Biol Chem. 2010 Dec 31;285(53):41244-54. doi: 10.1074/jbc.M110.155242. Epub 2010 Oct 29. J Biol Chem. 2010. PMID: 21036905 Free PMC article. - Functions and regulation of the Polo-like kinase Cdc5 in the absence and presence of DNA damage.
Botchkarev VV Jr, Haber JE. Botchkarev VV Jr, et al. Curr Genet. 2018 Feb;64(1):87-96. doi: 10.1007/s00294-017-0727-2. Epub 2017 Aug 2. Curr Genet. 2018. PMID: 28770345 Free PMC article. Review. - The function and regulation of budding yeast Swe1 in response to interrupted DNA synthesis.
Liu H, Wang Y. Liu H, et al. Mol Biol Cell. 2006 Jun;17(6):2746-56. doi: 10.1091/mbc.e05-11-1093. Epub 2006 Mar 29. Mol Biol Cell. 2006. PMID: 16571676 Free PMC article. - Tid1/Rdh54 translocase is phosphorylated through a Mec1- and Rad53-dependent manner in the presence of DSB lesions in budding yeast.
Ferrari M, Nachimuthu BT, Donnianni RA, Klein H, Pellicioli A. Ferrari M, et al. DNA Repair (Amst). 2013 May 1;12(5):347-55. doi: 10.1016/j.dnarep.2013.02.004. Epub 2013 Mar 7. DNA Repair (Amst). 2013. PMID: 23473644 Free PMC article.
References
- Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. - PMC - PubMed
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis resists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. - PubMed
- Charles J, Jaspersen S, Tinker-Kulberg R, Hwang L, Szidon A, Morgan D O. The polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. - PubMed
- Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases