Persistence of neutral polymorphisms in Lake Victoria cichlid fish - PubMed (original) (raw)
Persistence of neutral polymorphisms in Lake Victoria cichlid fish
S Nagl et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Phylogenetic trees for groups of closely related species often have different topologies, depending on the genes used. One explanation for the discordant topologies is the persistence of polymorphisms through the speciation phase, followed by differential fixation of alleles in the resulting species. The existence of transspecies polymorphisms has been documented for alleles maintained by balancing selection but not for neutral alleles. In the present study, transspecific persistence of neutral polymorphisms was tested in the endemic haplochromine species flock of Lake Victoria cichlid fish. Putative noncoding region polymorphisms were identified at four randomly selected nuclear loci and tested on a collection of 12 Lake Victoria species and their putative riverine ancestors. At all loci, the same polymorphism was found to be present in nearly all the tested species, both lacustrine and riverine. Different polymorphisms at these loci were found in cichlids of other East African lakes (Malawi and Tanganyika). The Lake Victoria polymorphisms must have therefore arisen after the flocks now inhabiting the three great lakes diverged from one another, but before the riverine ancestors of the Lake Victoria flock colonized the Lake. Calculations based on the mtDNA clock suggest that the polymorphisms have persisted for about 1.4 million years. To maintain neutral polymorphisms for such a long time, the population size must have remained large throughout the entire period.
Figures
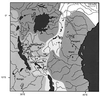
Figure 1
Geography of East Africa. Drainage systems are indicated by different shadings. A, Lake Victoria; B, Lake Malawi; C, Lake Tanganyika; D, Lake Manyara; E, Lake Chala; F, Lake Nabugabo; G, Lake Kayania; H, Lake Kayugi. Cichlids were caught at localities indicated by numbers.
Figure 2
Informative nucleotide sites of the loci HN49 and SN-Y. The top line shows the position of the nucleotide in the consensus sequence. A dash indicates identity to the consensus; an asterisk indicates a deletion.
Similar articles
- Phylogeny of African cichlid fishes as revealed by molecular markers.
Mayer WE, Tichy H, Klein J. Mayer WE, et al. Heredity (Edinb). 1998 Jun;80 ( Pt 6):702-14. doi: 10.1046/j.1365-2540.1998.00347.x. Heredity (Edinb). 1998. PMID: 9675872 - Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes.
Sturmbauer C, Baric S, Salzburger W, Rüber L, Verheyen E. Sturmbauer C, et al. Mol Biol Evol. 2001 Feb;18(2):144-54. doi: 10.1093/oxfordjournals.molbev.a003788. Mol Biol Evol. 2001. PMID: 11158373 - Mitochondrial phylogeny of the endemic mouthbrooding lineages of cichlid fishes from Lake Tanganyika in eastern Africa.
Sturmbauer C, Meyer A. Sturmbauer C, et al. Mol Biol Evol. 1993 Jul;10(4):751-68. doi: 10.1093/oxfordjournals.molbev.a040042. Mol Biol Evol. 1993. PMID: 8355599 - The taxonomic diversity of the cichlid fish fauna of ancient Lake Tanganyika, East Africa.
Ronco F, Büscher HH, Indermaur A, Salzburger W. Ronco F, et al. J Great Lakes Res. 2020 Oct;46(5):1067-1078. doi: 10.1016/j.jglr.2019.05.009. J Great Lakes Res. 2020. PMID: 33100489 Free PMC article. Review. - The species flocks of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics.
Salzburger W, Meyer A. Salzburger W, et al. Naturwissenschaften. 2004 Jun;91(6):277-90. doi: 10.1007/s00114-004-0528-6. Epub 2004 Apr 20. Naturwissenschaften. 2004. PMID: 15241604 Review.
Cited by
- Genetic structure of pelagic and littoral cichlid fishes from Lake Victoria.
Takeda M, Kusumi J, Mizoiri S, Aibara M, Mzighani SI, Sato T, Terai Y, Okada N, Tachida H. Takeda M, et al. PLoS One. 2013 Sep 6;8(9):e74088. doi: 10.1371/journal.pone.0074088. eCollection 2013. PLoS One. 2013. PMID: 24040175 Free PMC article. - Trans-Species Polymorphism in Immune Genes: General Pattern or MHC-Restricted Phenomenon?
Těšický M, Vinkler M. Těšický M, et al. J Immunol Res. 2015;2015:838035. doi: 10.1155/2015/838035. Epub 2015 May 24. J Immunol Res. 2015. PMID: 26090501 Free PMC article. Review. - SINEs as Credible Signs to Prove Common Ancestry in the Tree of Life: A Brief Review of Pioneering Case Studies in Retroposon Systematics.
Nikaido M, Nishihara H, Okada N. Nikaido M, et al. Genes (Basel). 2022 May 31;13(6):989. doi: 10.3390/genes13060989. Genes (Basel). 2022. PMID: 35741751 Free PMC article. Review. - Local Climate Heterogeneity Shapes Population Genetic Structure of Two Undifferentiated Insular Scutellaria Species.
Hsiung HY, Huang BH, Chang JT, Huang YM, Huang CW, Liao PC. Hsiung HY, et al. Front Plant Sci. 2017 Feb 10;8:159. doi: 10.3389/fpls.2017.00159. eCollection 2017. Front Plant Sci. 2017. PMID: 28239386 Free PMC article.
References
- Nei M. Am Nat. 1972;106:283–292.
- Pamilo P, Nei M. Mol Biol Evol. 1988;5:568–583. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources