Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms - PubMed (original) (raw)
Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms
Gro kopf R et al. Appl Environ Microbiol. 1998 Dec.
Abstract
Because excised, washed roots of rice (Oryza sativa) immediately produce CH4 when they are incubated under anoxic conditions (P. Frenzel and U. Bosse, FEMS Microbiol. Ecol. 21:25-36, 1996), we employed a culture-independent molecular approach to identify the methanogenic microbial community present on roots of rice plants. Archaeal small-subunit rRNA-encoding genes were amplified directly from total root DNA by PCR and then cloned. Thirty-two archaeal rice root (ARR) gene clones were randomly selected, and the amplified primary structures of ca. 750 nucleotide sequence positions were compared. Only 10 of the environmental sequences were affiliated with known methanogens; 5 were affiliated with Methanosarcina spp., and 5 were affiliated with Methanobacterium spp. The remaining 22 ARR gene clones formed four distinct lineages (rice clusters I through IV) which were not closely related to any known cultured member of the Archaea. Rice clusters I and II formed distinct clades within the phylogenetic radiation of the orders "Methanosarcinales" and Methanomicrobiales. Rice cluster I was novel, and rice cluster II was closely affiliated with environmental sequences obtained from bog peat in northern England. Rice cluster III occurred on the same branch as Thermoplasma acidophilum and marine group II but was only distantly related to these taxa. Rice cluster IV was a deep-branching crenarchaeotal assemblage that was closely related to clone pGrfC26, an environmental sequence recovered from a temperate marsh environment. The use of a domain-specific oligonucleotide probe in a fluorescent in situ hybridization analysis revealed that viable members of the Archaea were present on the surfaces of rice roots. In addition, we describe a novel euryarchaeotal main line of descent, designated rice cluster V, which was detected in anoxic rice paddy soil. These results indicate that there is an astonishing richness of archaeal diversity present on rice roots and in the surrounding paddy soil.
Figures
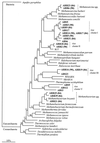
FIG. 1
Evolutionary distance dendrogram showing the positions of environmental SSU rDNA sequences recovered from rice roots (ARR sequences) and anoxic bulk soil (ABS sequences) from flooded rice microcosms. The positions of sequences are shown in relation to the positions of known members of the Euryarchaeota and environmental sequences retrieved from peat bogs (R17 [16]) and from coastal marine environments (WHAR N and SBAR 16 [9]). The numbers at the nodes indicate the percentages of recovery in 1,000 bootstrap resamplings. The numbers in parentheses indicate whether the environmental sequences were recovered from 84-day-old flooded rice microcosms or 90-day-old flooded rice microcosms. SSU rDNA sequences of Aquifex pyrophilus and of members of the Crenarchaeota were used as outgroup reference sequences. The tree topology was determined by using distance matrix methods (calculation of the distance matrix with the Jukes-Cantor equation [20], construction of the distance tree by the neighbor-joining method [31]). Scale bar = 10% difference in nucleotide sequence positions.
FIG. 2
Evolutionary distance dendrogram showing the positions of environmental SSU rDNA sequences recovered from rice roots (ARR sequences) and anoxic bulk soil (ABS sequences) from flooded rice microcosms. The positions of sequences are shown in relation to the positions of known members of the Crenarchaeota and environmental sequences retrieved from coastal marine environments (ANTARCTIC 12, SBAR5, and WHAR Q [9]), from a hot spring in the Yellowstone National Park (pJP27, pJP41, pJP78, and pJP 89 [4], as well as pSL12, pSL17, and pSL22 [3]), from shallow-sediment and marsh environments (pGrfC26 and pGrfA4 [17]), and from a forest soil (FFSB2 [21]). The numbers in parentheses indicate whether the environmental sequences were recovered from 84-day-old flooded rice microcosms or 90-day-old flooded rice microcosms. SSU rDNA sequences from Aquifex pyrophilus, from members of the Euryarchaeota, and from members of the “_Korarchaeota_” were used as outgroup reference sequences. The dendrogram was constructed as described in the legend to Fig. 1. Scale bar = 10% difference in nucleotide sequence positions.
FIG. 3
In situ detection of indigenous archaea on rice roots with rhodamine-labeled domain-specific oligonucleotide probe ARC915 (35). (A) The red dots in the center are coccoid archaeal cells that were 0.5 to 0.8 μm in diameter and specifically hybridized with probe ARC915. The photograph is an overlay resulting from three individual examinations, in situ hybridization with ARC915, DAPI staining, and autofluorescence of the plant tissue. The specificities of the hybridization signals were verified by measuring the relative signal intensities obtained from the oligonucleotide probe signal, the DAPI signal, and the autofluorescence signal, as shown for one optical cut in panel B (indicated by the arrow in panel A). The root cells are blue-green due to autofluorescence, and the dark areas correspond to the iron precipitates which often cover rice roots. The maximum distance between the archaeal cells and the root tissue was less than 12 μm, as determined by a z-series of optical sections, each 0.3 μm thick. Scale bar = 5 μm. (B) The three curves indicate the intensity of the oligonucleotide probe hybridization signal (red) in relation to the signal intensities of DAPI staining (blue) and autofluorescence of the plant tissue (green) for one optical cut with a high signal/noise ratio. The signal intensities were quantified by using the appropriate quantifying tools of the confocal laser scanning microscope.
Similar articles
- Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization(1).
Jurgens G, Glöckner F, Amann R, Saano A, Montonen L, Likolammi M, Münster U. Jurgens G, et al. FEMS Microbiol Ecol. 2000 Oct 1;34(1):45-56. doi: 10.1111/j.1574-6941.2000.tb00753.x. FEMS Microbiol Ecol. 2000. PMID: 11053735 - Methanogenic archaea and CO2-dependent methanogenesis on washed rice roots.
Lehmann-Richter S, Grosskopf R, Liesack W, Frenzel P, Conrad R. Lehmann-Richter S, et al. Environ Microbiol. 1999 Apr;1(2):159-66. doi: 10.1046/j.1462-2920.1999.00019.x. Environ Microbiol. 1999. PMID: 11207731 - Archaeal population dynamics during sequential reduction processes in rice field soil.
Lueders T, Friedrich M. Lueders T, et al. Appl Environ Microbiol. 2000 Jul;66(7):2732-42. doi: 10.1128/AEM.66.7.2732-2742.2000. Appl Environ Microbiol. 2000. PMID: 10877762 Free PMC article. - Rice Cluster I methanogens, an important group of Archaea producing greenhouse gas in soil.
Conrad R, Erkel C, Liesack W. Conrad R, et al. Curr Opin Biotechnol. 2006 Jun;17(3):262-7. doi: 10.1016/j.copbio.2006.04.002. Epub 2006 Apr 18. Curr Opin Biotechnol. 2006. PMID: 16621512 Review. - Microbiology of flooded rice paddies.
Liesack W, Schnell S, Revsbech NP. Liesack W, et al. FEMS Microbiol Rev. 2000 Dec;24(5):625-45. doi: 10.1111/j.1574-6976.2000.tb00563.x. FEMS Microbiol Rev. 2000. PMID: 11077155 Review.
Cited by
- Characterization of the bacterial and archaeal communities in rice field soils subjected to long-term fertilization practices.
Ahn JH, Song J, Kim BY, Kim MS, Joa JH, Weon HY. Ahn JH, et al. J Microbiol. 2012 Oct;50(5):754-65. doi: 10.1007/s12275-012-2409-6. Epub 2012 Nov 4. J Microbiol. 2012. PMID: 23124742 - "Methanoplasmatales," Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens.
Paul K, Nonoh JO, Mikulski L, Brune A. Paul K, et al. Appl Environ Microbiol. 2012 Dec;78(23):8245-53. doi: 10.1128/AEM.02193-12. Epub 2012 Sep 21. Appl Environ Microbiol. 2012. PMID: 23001661 Free PMC article. - Local conditions structure unique archaeal communities in the anoxic sediments of meromictic Lake Kivu.
Bhattarai S, Ross KA, Schmid M, Anselmetti FS, Bürgmann H. Bhattarai S, et al. Microb Ecol. 2012 Aug;64(2):291-310. doi: 10.1007/s00248-012-0034-x. Epub 2012 Mar 20. Microb Ecol. 2012. PMID: 22430505 - Phosphate inhibits acetotrophic methanogenesis on rice roots.
Conrad R, Klose M, Claus P. Conrad R, et al. Appl Environ Microbiol. 2000 Feb;66(2):828-31. doi: 10.1128/AEM.66.2.828-831.2000. Appl Environ Microbiol. 2000. PMID: 10653759 Free PMC article. - Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge.
Steinberg LM, Regan JM. Steinberg LM, et al. Appl Environ Microbiol. 2008 Nov;74(21):6663-71. doi: 10.1128/AEM.00553-08. Epub 2008 Sep 5. Appl Environ Microbiol. 2008. PMID: 18776026 Free PMC article.
References
- Aßmus B. In-situ-Detektion von Bakterien aus Boden- und Gewässerhabitaten mit spezifischen Markierungen sowie optischen und zytometrischen Methoden. Ph.D. thesis. Munich, Germany: Ludwig-Maximilians-Universität München; 1995.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous