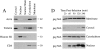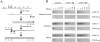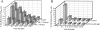Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton - PubMed (original) (raw)
Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton
A Bukrinskaya et al. J Exp Med. 1998.
Abstract
After interaction of human immunodeficiency virus type 1 (HIV-1) virions with cell surface receptors, a series of poorly characterized events results in establishment of a viral reverse transcription complex in the host cell cytoplasm. This process is coordinated in such a way that reverse transcription is initiated shortly after formation of the viral reverse transcription complex. However, the mechanism through which virus entry and initiation of reverse transcription are coordinated and how these events are compartmentalized in the infected cell are not known. In this study, we demonstrate that viral reverse transcription complexes associate rapidly with the host cell cytoskeleton during HIV-1 infection and that reverse transcription occurs almost entirely in the cytoskeletal compartment. Interruption of actin polymerization before virus infection reduced association of viral reverse transcription complexes with the cytoskeleton. In addition, efficient reverse transcription was dependent on intact actin microfilaments. The localization of reverse transcription to actin microfilaments was mediated by the interaction of a reverse transcription complex component (gag MA) with actin but not vimentin (intermediate filaments) or tubulin (microtubules). In addition, fusion, but not endocytosis-mediated HIV-1 infectivity, was impaired when actin depolymerizing agents were added to target cells before infection but not when added after infection. These results point to a previously unsuspected role for the host cell cytoskeleton in HIV-1 entry and suggest that components of the cytoskeleton promote establishment of the reverse transcription complex in the host cell and also the process of reverse transcription within this complex.
Figures
Figure 1
Association of viral reverse transcription complexes with the cytoskeleton. MT-4 cells were infected with HIV-1, and at 1 h after infection, cells were fractionated as detailed in Materials and Methods (sizes to the right in kD). Subcellular fractions were examined for the presence of cellular proteins (A) and virion-derived gag MA protein (B). The distribution of phosphorylated (Phospho.) gag MA, which specifically associates with the reverse transcription complex, is shown in C. MT-4 cells were fractionated at various intervals after HIV-1 infection, and the distribution of gag MA in various cellular compartments was determined by Western blotting (D). All Western blot images are within the linear range of the ECL assay. For A, B, and C, cell lysate from a total of 106 cells was loaded in each lane.
Figure 1
Association of viral reverse transcription complexes with the cytoskeleton. MT-4 cells were infected with HIV-1, and at 1 h after infection, cells were fractionated as detailed in Materials and Methods (sizes to the right in kD). Subcellular fractions were examined for the presence of cellular proteins (A) and virion-derived gag MA protein (B). The distribution of phosphorylated (Phospho.) gag MA, which specifically associates with the reverse transcription complex, is shown in C. MT-4 cells were fractionated at various intervals after HIV-1 infection, and the distribution of gag MA in various cellular compartments was determined by Western blotting (D). All Western blot images are within the linear range of the ECL assay. For A, B, and C, cell lysate from a total of 106 cells was loaded in each lane.
Figure 2
Inhibitors of actin polymerization reduce cytoskeletal association of viral reverse transcription complexes. (A) MT-4 cells were treated for 2 h with CCD (5 μM), which prevents actin polymerization. Cells were then infected with HIV-1 and fractionated at 1 and 5 h after infection (P.I.) for analysis of gag MA distribution. (B) MT-4 cells were treated with nocodazole (10 μM), an inhibitor of tubulin polymerization into microtubules, or demecolcine (10 μM), which causes disaggregation of intermediate filaments. Gag MA in the cytoskeleton was examined by Western blotting 1 h after infection. (C) MT-4 cells were incubated with CCD for 1 and 2 h before infection or 1 and 2 h after infection. At 3 h after infection, cytoskeletal and nuclear extracts were prepared and examined for the presence of gag MA by Western blotting. (D) The subcellular distribution of actin was examined by Western blotting in MT-4 cells (Control) and in MT-4 cells after a 2-h incubation with CCD. For A, B, and C, equal amounts of cellular protein were loaded in each lane. In D, each lane contains lysate from 106 cells.
Figure 3
Interaction between cytoskeletal proteins and HIV-1 gag MA in vivo. (A) Cell lysates of HIV-1–infected and uninfected MT-4 cells were immunoprecipitated with antibodies to gag MA and gag CA in 100, 300, and 800 mM NaCl. The presence of viral and cytoskeletal proteins in gag MA and gag CA immune complexes was examined by Western blotting (B) with antibodies to viral (gag MA, gag CA) and cytoskeletal (actin, tubulin, vimentin) proteins. (C) Western blot analysis of cytoskeletal proteins in cell lysates at different NaCl concentrations.
Figure 4
Efficient HIV-1 reverse transcription requires actin microfilaments. (A) Schematic of the major steps involved in synthesis of HIV-1 cDNA. Broken lines, genomic RNA; solid lines, cDNA; short arrows, regions of the viral genome to which PCR primers are directed; asterisk, the primer binding site. R-U5 and U5-gag primers amplify predominantly early and late products of reverse transcription, respectively. (B) HIV-1–infected MT-4 cells were incubated in the presence or absence of CCD (5 μM, added 1 h before or 2 h after HIV-1 infection), and subcellular fractions were prepared at 3 h after infection. Lysates were treated with DNAse to remove carryover DNA in the inoculum (reference 37). Early (LTR R-U5) and late (LTR U5-gag) products of reverse transcription in cell fractions were quantitated by PCR in serial fivefold dilutions of the cellular lysate. Infections carried out in the presence of AZT (5 μM) were used to verify de novo synthesis of viral cDNA. PCR products were visualized by Southern blot hybridization to HIV-1–specific oligonucleotide probes as outlined in Materials and Methods.
Figure 5
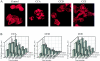
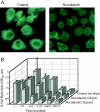
Actin microfilaments are required for efficient HIV-1 infection. (A) Immunohistochemical analysis of HeLa-CD4-LTR/β-gal indicator cells with rhodamine-phalloidin after a 1-h incubation with the actin depolymerizing agents CCA, CCD, and CCE (5 μM). In untreated (Control) cells, actin filaments are organized in linear array stress fibers of polymerized actin. Cytochalasin (5 μM) was sufficient to almost completely prevent actin polymerization in these cells. (B) Magi cells were incubated with the indicated cytochalasins for 2 h before (2h pre) and 2 h after (2h post) virus infection. Infected cells were harvested 48 h later for quantitation of β-galactosidase production. Cell infection was evaluated at five different levels of input virus in triplicate wells.
Figure 6
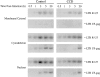
Efficient synthesis of late reverse transcription products requires actin microfilaments. Untreated (Control) and CCD-treated Magi cells (5 μM CCD added 2 h before virus infection) were harvested at different intervals after infection, and early and late products of reverse transcription in different subcellular compartments were examined by PCR using LTR R-U5 and LTR U5-gag primers, respectively.
Figure 7
HIV-1 infection does not require microtubules. (A) Organization of microtubules in Magi cells after incubation (2 h) with nocodazole, a microtubule depolymerizing drug that fixes monomers of tubulin. (B) Single-cycle HIV-1 infectivity characteristics in untreated and nocodazole-treated Magi cells. Infectivity was analyzed in triplicate at six different levels of input virus. Effects of nocodazole on single-cycle HIV-1 infection of Magi cells are as outlined in the legend to Fig. 5.
Figure 8
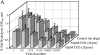
Inhibitors of myosin light-chain kinase impair HIV-1 infectivity. Single-cycle infectivity analysis of HIV-1 was examined in Magi cells treated with myosin light-chain kinase inhibitors ML-7 (20 μM) and KT5926 (A and B, respectively). (C) Gag MA distribution in cytosol/ membrane, cytoskeletal, and nuclear extracts of Magi cells after treatment with the indicated concentrations of KT5926 was examined by Western blotting. Relative amounts of gag MA in each sample were determined by volume integration (ImageQuant software; Molecular Dynamics).
Figure 8
Inhibitors of myosin light-chain kinase impair HIV-1 infectivity. Single-cycle infectivity analysis of HIV-1 was examined in Magi cells treated with myosin light-chain kinase inhibitors ML-7 (20 μM) and KT5926 (A and B, respectively). (C) Gag MA distribution in cytosol/ membrane, cytoskeletal, and nuclear extracts of Magi cells after treatment with the indicated concentrations of KT5926 was examined by Western blotting. Relative amounts of gag MA in each sample were determined by volume integration (ImageQuant software; Molecular Dynamics).
Figure 9
Actin microfilaments are not required for HIV-1 infection via endocytosis. (A) Effect of CCD on single-cycle infectivity of VSV-G– pseudotyped HIV-1. Virions containing VSV envelope proteins were obtained after cotransfection of an HIV-1 envelope minus mutant with VSV-G expression vector. Pseudotyped virions were obtained from transfected cell supernatants, and infectivities were determined in CCD-treated and untreated Magi cells. (B) Untreated (Control) and CCD-treated (10 μM CCD added 2 h before infection) Magi cells were infected with VSV-G– pseudotyped HIV-1 virions. Cytoskeletal and nuclear extracts were prepared 4 h after infection for analysis of gag MA distribution by Western blotting.
Figure 9
Actin microfilaments are not required for HIV-1 infection via endocytosis. (A) Effect of CCD on single-cycle infectivity of VSV-G– pseudotyped HIV-1. Virions containing VSV envelope proteins were obtained after cotransfection of an HIV-1 envelope minus mutant with VSV-G expression vector. Pseudotyped virions were obtained from transfected cell supernatants, and infectivities were determined in CCD-treated and untreated Magi cells. (B) Untreated (Control) and CCD-treated (10 μM CCD added 2 h before infection) Magi cells were infected with VSV-G– pseudotyped HIV-1 virions. Cytoskeletal and nuclear extracts were prepared 4 h after infection for analysis of gag MA distribution by Western blotting.
Similar articles
- Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1.
Jolly C, Mitar I, Sattentau QJ. Jolly C, et al. J Virol. 2007 Jun;81(11):5547-60. doi: 10.1128/JVI.01469-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360745 Free PMC article. - HIV-1 Exploits a Dynamic Multi-aminoacyl-tRNA Synthetase Complex To Enhance Viral Replication.
Duchon AA, St Gelais C, Titkemeier N, Hatterschide J, Wu L, Musier-Forsyth K. Duchon AA, et al. J Virol. 2017 Oct 13;91(21):e01240-17. doi: 10.1128/JVI.01240-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28814526 Free PMC article. - Cellular minichromosome maintenance complex component 5 (MCM5) is incorporated into HIV-1 virions and modulates viral replication in the newly infected cells.
Santos S, Obukhov Y, Nekhai S, Pushkarsky T, Brichacek B, Bukrinsky M, Iordanskiy S. Santos S, et al. Virology. 2016 Oct;497:11-22. doi: 10.1016/j.virol.2016.06.023. Epub 2016 Jul 12. Virology. 2016. PMID: 27414250 Free PMC article. - The host cytoskeleton: a key regulator of early HIV-1 infection.
Stephens C, Naghavi MH. Stephens C, et al. FEBS J. 2024 May;291(9):1835-1848. doi: 10.1111/febs.16706. Epub 2022 Dec 26. FEBS J. 2024. PMID: 36527282 Review. - The trinity of the cortical actin in the initiation of HIV-1 infection.
Spear M, Guo J, Wu Y. Spear M, et al. Retrovirology. 2012 May 28;9:45. doi: 10.1186/1742-4690-9-45. Retrovirology. 2012. PMID: 22640593 Free PMC article. Review.
Cited by
- Help or Hinder: Protein Host Factors That Impact HIV-1 Replication.
Moezpoor MR, Stevenson M. Moezpoor MR, et al. Viruses. 2024 Aug 10;16(8):1281. doi: 10.3390/v16081281. Viruses. 2024. PMID: 39205255 Free PMC article. Review. - CD147 stimulates HIV-1 infection in a signal-independent fashion.
Pushkarsky T, Yurchenko V, Laborico A, Bukrinsky M. Pushkarsky T, et al. Biochem Biophys Res Commun. 2007 Nov 23;363(3):495-9. doi: 10.1016/j.bbrc.2007.08.192. Epub 2007 Sep 14. Biochem Biophys Res Commun. 2007. PMID: 17888876 Free PMC article. - Proteomic analysis of primary duck hepatocytes infected with duck hepatitis B virus.
Zhao Y, Ben H, Qu S, Zhou X, Yan L, Xu B, Zhou S, Lou Q, Ye R, Zhou T, Yang P, Qu D. Zhao Y, et al. Proteome Sci. 2010 Jun 7;8:28. doi: 10.1186/1477-5956-8-28. Proteome Sci. 2010. PMID: 20529248 Free PMC article. - Chemokine coreceptor signaling in HIV-1 infection and pathogenesis.
Wu Y, Yoder A. Wu Y, et al. PLoS Pathog. 2009 Dec;5(12):e1000520. doi: 10.1371/journal.ppat.1000520. Epub 2009 Dec 24. PLoS Pathog. 2009. PMID: 20041213 Free PMC article. Review. - Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells.
Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. Naghavi MH, et al. EMBO J. 2007 Jan 10;26(1):41-52. doi: 10.1038/sj.emboj.7601475. Epub 2006 Dec 14. EMBO J. 2007. PMID: 17170707 Free PMC article.
References
- Dragic, T., A. Trkola, and J.P. Moore. 1997. HIV co-receptors: gateways to the cell. In HIV Advances in Research and Therapy. Vol. 7. T.C. Merigan, D.P. Bolognesi, J. Feinberg, P.A. Volberding, and F. Wong-Staal, editors. Cliggott Communications, Greenwich, CT. 3–12.
- Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9:817–823. - PubMed
- Heinzinger N, Bukrinsky M, Haggerty S, Ragland A, Lee M-A, Kewalramani V, Gendelman H, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical