Functional characterization of a Nup159p-containing nuclear pore subcomplex - PubMed (original) (raw)
Functional characterization of a Nup159p-containing nuclear pore subcomplex
N Belgareh et al. Mol Biol Cell. 1998 Dec.
Free PMC article
Abstract
Nup159p/Rat7p is an essential FG repeat-containing nucleoporin localized at the cytoplasmic face of the nuclear pore complex (NPC) and involved in poly(A)+ RNA export and NPC distribution. A detailed structural-functional analysis of this nucleoporin previously demonstrated that Nup159p is anchored within the NPC through its essential carboxyl-terminal domain. In this study, we demonstrate that Nup159p specifically interacts through this domain with both Nsp1p and Nup82p. Further analysis of the interactions within the Nup159p/Nsp1p/Nup82p subcomplex using the nup82Delta108 mutant strain revealed that a deletion within the carboxyl-terminal domain of Nup82p prevents its interaction with Nsp1p but does not affect the interaction between Nup159p and Nsp1p. Moreover, immunofluorescence analysis demonstrated that Nup159p is delocalized from the NPC in nup82Delta108 cells grown at 37 degrees C, a temperature at which the Nup82Delta108p mutant protein becomes degraded. This suggests that Nup82p may act as a docking site for a core complex composed of the repeat-containing nucleoporins Nup159p and Nsp1p. In vivo transport assays further revealed that nup82Delta108 and nup159-1/rat7-1 mutant strains have little if any defect in nuclear protein import and protein export. Together our data suggest that the poly(A)+ RNA export defect previously observed in nup82 mutant cells might be due to the loss from the NPCs of the repeat-containing nucleoporin Nup159p.
Figures
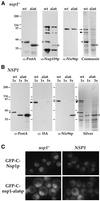
Figure 1
Nup159p interacts with Nup82p and Nsp1p. (A and B) ProtA-Nup49p, ProtA-Nup82p, and ProtA-C-Nsp1p expressed in the protease-deficient BJ2168 strain were affinity purified by chromatography on IgG-Sepharose as described in MATERIALS AND METHODS. (A) Purified fractions from the ProtA-Nup49p, ProtA-C-Nsp1p, and ProtA-Nup82p affinity columns were analyzed by immunoblotting after SDS-PAGE, using IgG coupled to HRP to visualize the ProtA-fusion proteins (α-ProtA) and an anti-Nup159p antibody directed against the repeat motifs of Nup159p (rat7#4, see MATERIALS AND METHODS). The positions of the ProtA fusion proteins (dots) are indicated. Note that the rat7#4 antibody also cross-reacts with ProtA chimeras. (B) Eluted fractions from the ProtA-Nup49p and ProtA-Nup82p affinity columns were analyzed by SDS-PAGE followed by Coomassie blue staining or by Western blotting using IgG coupled to HRP to visualize the ProtA constructs, an antibody directed against the carboxyl-terminal domain of Nup159p (rat7#5), and an anti-Nsp1p antibody directed against the repeat domain of Nsp1p. Coomassie blue staining reveals Nsp1p, Nic96p, and Nup57p in the ProtA-Nup49p–purified fraction, whereas Nsp1p and Nup159p copurify with the ProtA-Nup82p fusion protein. The positions of Nup159p, Nsp1p, Nic96p, Nup57p (arrows), and the ProtA fusion proteins (dots) are indicated. (C) Affinity purification of ProtA-C-Nsp1p expressed in an _NSP1_-disrupted strain. Equivalent amounts of the load (soluble fraction from the whole-cell lysates [S]), of the unbound (flow-through [F]), and of the bound and subsequently eluted (E) fractions were analyzed by Western blotting. IgGs coupled to HRP were used to detect ProtA-C-Nsp1p, the rat7#5 antibody to visualize Nup159p, and antibodies directed against Nic96p and Nup133p were used as controls. Note that the _NSP1_-disrupted strain is not protease deficient, leading to a substantial degradation of Nup159p with a major degradation product migrating at ∼100 kDa.
Figure 2
In the presence of wild-type Nsp1p, Nsp1-ala6p no longer interacts with Nup82p and Nic96p and is not targeted to the NPC. (A) Yeast cells with a disrupted chromosomal copy of NSP1 and complemented by ProtA-C-Nsp1p (wt) or ProtA-C-nsp1-ala6p (ala6) were grown at 24°C. Whole-cell lysates from these two strains were affinity purified by IgG-Sepharose chromatography. The affinity-purified fractions were analyzed by Western blotting for the presence of ProtA-C-Nsp1p (asterisks) or ProtA-C-nsp1-ala6p (open circles), Nup159p (using the rat7#5 antibody), and Nic96p, or by Coomassie blue staining. In this _nsp1_− background, both ProtA-C-Nsp1p and ProtA-C-nsp1-ala6p were able to interact with Nup159p and Nic96p. Nic96p (arrow) and Nup82p (arrowhead) could also be detected by Coomassie blue staining of the two eluates. Note that in this strain background, Nup159p was degraded during the purification procedure and was therefore hardly detectable by Coomassie blue staining. (B) Whole-cell lysates from NUP82-HA cells carrying a wild-type chromosomal copy of NSP1 and expressing ProtA-C-Nsp1p (wt) or ProtA-C-nsp1-ala6p (ala6) were affinity purified byIgG-Sepharose chromatography as described in MATERIALS AND METHODS. The eluates were analyzed by silver staining (Silver) or by Western blot using an anti-IgG coupled to HRP to detect ProtA-C-Nsp1p (asterisks) or ProtA-C-nsp1-ala6p (open circles), an anti-HA antibody to detect Nup82-HAp (arrrowhead), or an antiNic96p (arrow). In the presence of wild-type Nsp1p, the interaction between ProtA-C-nsp1-ala6p and both Nup82-HAp and Nic96p was inhibited (a very faint band was detectable only when a fivefold excess of the ProtA-C-nsp1-ala6 eluate was loaded). (C) The GFP-C-NSP1 and GFP-C-nsp1-ala6 fusion genes were expressed in an _NSP1-_disrupted strain (_nsp1_−) and in a wild-type strain (NSP1). The GFP-C-Nsp1p and GFP-C-nsp1-ala6p signals were observed by fluorescence microscopy in living cells grown at 24°C as described in MATERIALS AND METHODS.
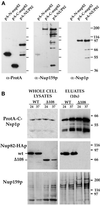
Figure 3
Nsp1p and Nup159p form a core complex independent of interaction with the carboxyl-terminal domain of Nup82p. (A) Affinity purification by IgG-Sepharose chromatography of ProtA-N-nup82p, ProtA-C-nup82p, and ProtA-Nup82p expressed in strains derived from BJ2168. The purified fractions were analyzed by Western blotting using IgG coupled to HRP to detect the ProtA fusions, an anti-Nup159p antibody directed against the carboxyl-terminal domain of Nup159p (rat7#5), and an antibody directed against the repeat domain of Nsp1p. (B) Whole-cell lysates from NUP82 (WT) and nup82-Δ108 (Δ108) strains transformed with the pRS316-ProtA-C-Nsp1p plasmid and maintained at 24°C or shifted to 37°C for 3 h were affinity purified by IgG-Sepharose chromatography. Whole-cell lysates and affinity-purified fractions (eluates, 10-fold equivalent) were analyzed by Western blotting for the presence of ProtA-C-Nsp1p using an anti-IgG coupled to HRP, for the presence of Nup82-HAp and Nup82-Δ108-HAp with a monoclonal antibody directed against the HA epitope, and for the presence ofNup159p with the rat7#4 antibody. Although Nup82-Δ108-HAp does not interact with ProtA-C-Nsp1p at either 24 or 37°C, Nup159p still copurifies with ProtA-C-Nsp1p in the nup82Δ108 strain. Note that the NUP82-wt and nup82Δ108 strains are not protease deficient, leading to a substantial degradation of Nup159p. The shorter degradation product of Nup159p did not copurify with ProtA-C-Nsp1p.
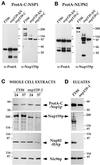
Figure 4
A deletion within the carboxyl-terminal domain of Nup159p impairs the stability of the Nup159p/Nsp1p/Nup82p complex. (A and B) ProtA-C-Nsp1p (A) or ProtA-Nup82p (B), expressed in FY86 (NUP159-wt), a nup159ΔN, and a nup159-C strain were affinity purified using IgG-Sepharose chromatography, and the affinity-purified fractions were analyzed by Western blotting. IgG coupled to HRP was used to detect the ProtA fusion proteins, whereas the presence of Nup159p was detected using the rat7#5 antibody directed against the carboxyl-terminal domain of Nup159p. Note that full-length Nup159p and Nup159ΔNp proteins were highly sensitive to proteolysis, leading to several breakdown products recognized by the anti-Nup159p antibody, whereas no proteolysis of the Nup159-Cp protein was observed. (C) FY86 (wt) and nup159-1 strains, transformed with the pRS316-ProtA-C-NSP1 and pRS314-Nup82-HA plasmids, were grown at 24°C or shifted to 37°C for 3 h. Whole-cell extracts were prepared as described in MATERIALS AND METHODS and analyzed by Western blotting for the presence of ProtA-C-Nsp1p, Nup82-HAp, Nup159p (using the rat7#4 antibody that presents the same affinity for Nup159p and for the truncated Nup159-1p), and Nic96p. After a 3-h shift to 37°C, which induces the degradation of Nup159-1p, the apparent expression level of Nup82-HAp is also decreased. (D) Whole-cell lysates from these two strains grown at 24°C were affinity purified by IgG-Sepharose chromatography. The affinity-purified fractions (eluates) were analyzed by Western blotting and tested for the presence of ProtA-C-Nsp1p, Nup82-HAp, Nup159p, and Nic96p. Unlike wild-type Nup159p, Nup159-1p does not copurify with ProtA-C-Nsp1p. In this mutant strain, the interaction between Nup82-HAp and ProtA-C-Nsp1p was also inhibited, whereas Nic96p still copurified with ProtA-C-Nsp1p.
Figure 5
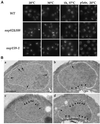
Nup159p is lost from the NPCs when nup159-1 or nup82Δ108 cells are shifted to 37°C. Wild-type (NUP82-wt), nup159-1, nup82Δ108, nup82Δ87, and ala6-nsp1 cells were grown at 23°C or shifted for 3 h to 37°C. The subcellular localization of Nup159p was analyzed by immunofluorescence using a polyclonal antibody directed against the repeat-containing domain of Nup159p. Cells were also stained for DNA with DAPI. At 23°C, nuclear pore labeling was observed in all five strains. After a shift to 37°C, a wild-type staining pattern was still observed in the NUP82-wt, nup82Δ87, and ala6-nsp1 strains, whereas no perinuclear staining could be detected in the nup159-1 and nup82Δ108 strains, indicating that Nup159p had been delocalized from the NPCs in these two strains.
Figure 6
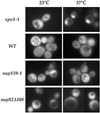
In vivo localization of GFP-Nup82p and GFP-Nsp1p in the nup159-1 or nup82Δ108 mutant strains. (A) The GFP-NUP82 fusion gene was expressed in a _NUP82_-disrupted strain and in a nup82::HIS3/nup159-1 strain (strain YV384). The GFP-Nup82p staining was observed by fluorescence microscopy in living cells grown at 20°C or shifted for 3 h to 37°C as described in MATERIALS AND METHODS. In wild-type NUP159 cells, the GFP-Nup82p fusion protein was localized at the NPCs and gave a ring-like staining at both 20 and 37°C. In nup159-1 mutant cells grown at 20°C, the GFP-Nup82p was less homogeneously distributed around the nuclear envelope because of the NPC clustering phenotype of the nup159-1 strain. After a 3-h shift to 37°C that leads to a redistribution of the NPCs, the GFP-Nup82p was still localized at the NPC in the nup159-1 strain, although enhanced cytoplasmic staining is also evident. (B) The GFP-C-NSP1 construct was introduced in a wild-type (WT) strain and the nup159-1 and nup82Δ108 strains. Cells were grown at 20°C or shifted for 3 h at 37°C. In wild-type cells, GFP-Nsp1p gave a ring-like staining at both 20 and 37°C. In nup159-1 and nup82Δ108 strains, the GFP-Nsp1p labeling was restricted to a few clusters at 20°C and gave rise to a homogeneous ring staining after a 3-h shift to 37°C.
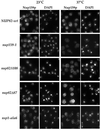
Figure 7
The nup82Δ108 mutant exhibits a mild and reversible NPC clustering phenotype. (A) To analyze NPC distribution in vivo, wild-type (WT), nup82Δ108, and nup159-1 strains were transformed with the pFL38-GFP-NUP49 plasmid. Yeast cells were grown in liquid SD medium lacking uracil to early log phase at 20 or 30°C, grown at 20°C, and shifted for 1 h to 37°C or grown on plates at 20°C for 2 d. These cells were examined for GFP fluorescence as described in MATERIALS AND METHODS. Wild-type cells displayed a homogeneous labeling pattern around the nuclear envelope at all temperatures. In contrast, a mild clustering phenotype was observed in nup82Δ108 cells and nup159-1 cells at 20°C and to a lesser extent at 30°C. This clustering phenotype is reversed after a 1-hour shift to 37°C. Strikingly, nup82Δ108 and nup159-1 cells grown on plates at 20°C exhibited a homogeneous staining of the nuclear envelope. (B) Electron micrographs of nuclei from wild-type (a) and nup82Δ108 (b–d) strains grown at 16°C. Wild-type cells have nuclei with evenly distributed NPCs, whereas nup82Δ108 cells exhibited grouped or clustered NPCs. Bar, 0.5 μm. Arrows point to single NPCs, and arrowheads point to clustered nuclear pores.
Figure 8
nup159-1 and nup82Δ108 cells do not display a major defect in NES-mediated protein export at 37°C. xpo1-1, wild-type (FY86), nup159-1, and nup82Δ108 cells were transformed with the NES-GFP2-NLS plasmid. Cells were maintained at 23°C or shifted to 37°C for 3 h. In xpo1-1 cells shifted to 37°C, an intranuclear accumulation of the NES-GFP2-NLS reporter and a concomitant decrease of the cytoplasmic staining was observed. In contrast, only a slight perinuclear accumulation of the reporter fusion protein could be observed in nup159-1 and nup82Δ108 cells shifted to 37°C.
Figure 9
In vivo analysis of the nuclear import of Mig1-GFP-lacZ in ts nup159, nsp1, and nup82 mutant strains. Temperature-sensitive nup159, nsp1, and nup82 mutant cells expressing the Mig1-GFP-LacZ reporter protein were grown in glycerol-containing medium at the indicated temperatures. At this stage, glucose was added, and living cells were examined and recorded at 1, 3, 5, and 8 min after glucose addition. A rapid nuclear import of the reporter was observed in the nup159-1 and nup82Δ108 mutant strains at both 23 and 37°C. In contrast, a modest defect in import was seen in nsp1-ala6 mutant cells grown at 25°C, and only a few nsp1-ala6 mutant cells shifted for 2 h to 37°C displayed any import of reporter protein, even 8 min after glucose addition. Note that to improve the detection of the faint nuclear signal, the images have been converted to negative images using the Adobe Photoshop program.
Similar articles
- Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex.
Bailer SM, Balduf C, Katahira J, Podtelejnikov A, Rollenhagen C, Mann M, Pante N, Hurt E. Bailer SM, et al. J Biol Chem. 2000 Aug 4;275(31):23540-8. doi: 10.1074/jbc.M001963200. J Biol Chem. 2000. PMID: 10801828 - Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p.
Ho AK, Shen TX, Ryan KJ, Kiseleva E, Levy MA, Allen TD, Wente SR. Ho AK, et al. Mol Cell Biol. 2000 Aug;20(15):5736-48. doi: 10.1128/MCB.20.15.5736-5748.2000. Mol Cell Biol. 2000. PMID: 10891509 Free PMC article. - The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport.
Bailer SM, Balduf C, Hurt E. Bailer SM, et al. Mol Cell Biol. 2001 Dec;21(23):7944-55. doi: 10.1128/MCB.21.23.7944-7955.2001. Mol Cell Biol. 2001. PMID: 11689687 Free PMC article. - A structure/function analysis of Rat7p/Nup159p, an essential nucleoporin of Saccharomyces cerevisiae.
Del Priore V, Heath C, Snay C, MacMillan A, Gorsch L, Dagher S, Cole C. Del Priore V, et al. J Cell Sci. 1997 Dec;110 ( Pt 23):2987-99. doi: 10.1242/jcs.110.23.2987. J Cell Sci. 1997. PMID: 9359887 - The nuclear pore complex.
Davis LI. Davis LI. Annu Rev Biochem. 1995;64:865-96. doi: 10.1146/annurev.bi.64.070195.004245. Annu Rev Biochem. 1995. PMID: 7574503 Review.
Cited by
- RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase.
Askjaer P, Bachi A, Wilm M, Bischoff FR, Weeks DL, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj IW, Fornerod M. Askjaer P, et al. Mol Cell Biol. 1999 Sep;19(9):6276-85. doi: 10.1128/MCB.19.9.6276. Mol Cell Biol. 1999. PMID: 10454574 Free PMC article. - Fantastic nuclear envelope herniations and where to find them.
Thaller DJ, Patrick Lusk C. Thaller DJ, et al. Biochem Soc Trans. 2018 Aug 20;46(4):877-889. doi: 10.1042/BST20170442. Epub 2018 Jul 19. Biochem Soc Trans. 2018. PMID: 30026368 Free PMC article. Review. - Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution.
Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Chadrin A, et al. J Cell Biol. 2010 May 31;189(5):795-811. doi: 10.1083/jcb.200910043. Epub 2010 May 24. J Cell Biol. 2010. PMID: 20498018 Free PMC article. - The Great Escape: mRNA Export through the Nuclear Pore Complex.
De Magistris P. De Magistris P. Int J Mol Sci. 2021 Oct 29;22(21):11767. doi: 10.3390/ijms222111767. Int J Mol Sci. 2021. PMID: 34769195 Free PMC article. Review. - The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution.
Ulrich A, Partridge JR, Schwartz TU. Ulrich A, et al. Mol Biol Cell. 2014 May;25(9):1484-92. doi: 10.1091/mbc.E13-12-0745. Epub 2014 Feb 26. Mol Biol Cell. 2014. PMID: 24574455 Free PMC article.
References
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases