Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression - PubMed (original) (raw)
doi: 10.1128/JVI.73.1.161-169.1999.
P Sharma, L Dudus, Y Zhang, S Sanlioglu, Z Yan, Y Yue, Y Ye, R Lester, J Yang, K J Fisher, J F Engelhardt
Affiliations
- PMID: 9847318
- PMCID: PMC103819
- DOI: 10.1128/JVI.73.1.161-169.1999
Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression
D Duan et al. J Virol. 1999 Jan.
Abstract
A central feature of the adeno-associated virus (AAV) latent life cycle is persistence in the form of both integrated and episomal genomes. However, the molecular processes associated with episomal long-term persistence of AAV genomes are only poorly understood. To investigate these mechanisms, we have utilized a recombinant AAV (rAAV) shuttle vector to identify circular AAV intermediates from transduced HeLa cells and primary fibroblasts. The unique structural features exhibited by these transduction intermediates included circularized monomer and dimer virus genomes in a head-to-tail array, with associated specific base pair alterations in the 5' viral D sequence. In HeLa cells, the abundance and stability of AAV circular intermediates were augmented by adenovirus expressing the E2a gene product. In the absence of E2a, adenovirus expressing the E4 open reading frame 6 gene product decreased the abundance of AAV circular intermediates, favoring instead the linear replication form monomer (Rfm) and dimer (Rfd) structures. In summary, the formation of AAV circular intermediates appears to represent a new pathway for AAV genome conversion, which is consistent with the head-to-tail concatemerization associated with latent-phase persistence of rAAV. A better understanding of this pathway may increase the utility of rAAV vectors for gene therapy.
Figures
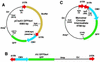
FIG. 1
Structure of the provirus shuttle vector and the predicted structure of rAAV circular intermediate monomers. With the aid of an rAAV _cis_-acting plasmid, pCisAV.GFP3ori (A), we produced the AV.GFP3ori recombinant virus (B). This vector encoded a GFP transgene cassette, an Ampr gene, and a bacterial replication origin (Ori). The predominant form of circular intermediates isolated following transduction of HeLa cells with AV.GFP3ori consisted of head-to-tail monomers (C).
FIG. 2
Structural analysis of rAAV circular intermediates in HeLa cells. Circular rAAV intermediate clones isolated from AV.GFP3ori-infected HeLa cells were analyzed by diagnostic restriction digestion with _Ase_I, _Sph_I, and _Pst_I, together with Southern blotting against ITR, GFP, and stuffer 32P-labeled probes (only data for ITR probes are shown). In panel A, four clones representing the diversity of intermediates found (p190, p333, p280, and p345) gave a diagnostic _Pst_I (P) restriction pattern (3- and 1.7-kb bands) consistent with a circular monomer or multimer intact genome (agarose gel [left] and Southern blot [right]). _Sph_I (S) digestion demonstrated the existence of a single ITR (p190), two ITRs in a head-to-tail orientation (p333 and p280), or three ITRs (p345) in isolated circular intermediates. The restriction patterns of pCisAV.GFP3ori (U, uncut; P, _Pst_I cut; S, _Sph_I cut) and a 1-kb DNA ladder (L) are also given for comparison. One additional circular form (p340) that was repetitively seen had an unidentifiable structure which lacked intact ITR sequences. Circular concatemers were identified by partial digestion with _Ase_I for clones p280 (dimer) and p333 (monomer), as is shown in panel B. Sequence analysis (C) of six clones with restriction patterns identical to that of p333 (A) was performed with primers (indicated by arrows) juxtaposed with the partial AAV 3′-polyadenylation sequences (dotted line) which flank the pSub201-derived ITRs (solid line). The top sequence represents the proposed head-to-tail structure of intact ITR arrays with sequence alignment derived from individual clones. Only partial sequence was achievable, due to the high secondary structure of ITRs (unknown sequence is marked by dashes in the sequence alignment). The junction of the inverted ITRs is marked by inverted arrowheads (at 251 bp). Several consistent base pair changes (shaded) were noted in the 5′ ITR D sequence (boxed) within four clones (p79, p81, p87, and p88). All base pair changes are indicated in lowercase letters.
FIG. 3
Adenovirus augments AAV circular intermediate formation in HeLa cells. Infection of HeLa cells with increasing doses (0, 500, and 5,000 particles/cell) of recombinant E1-deleted adenovirus (Ad.CMVlacZ) led to substantial expression of E2a 72-kDa DBP, as demonstrated by indirect immunofluorescent staining for DBP at 72 h postinfection (A). Coinfection of HeLa cells with Ad.CMVlacZ (5,000 particles/cell) and AV.GFP3ori (1,000 DNA particles/cell) led to substantial augmentation of rAAV GFP transgene expression (B). Augmentation of rAAV GFP transgene expression in the presence of increasing amounts (0, 500, 5,000, and 10,000 particles/cell) of recombinant Ad.CMVlacZ was quantified by FACS analysis at 72 h postinfection (C). Results demonstrate the mean (± standard error) for two experiments performed in duplicate. In addition, an aliquot of cells was split (1:10) at the time of FACS analysis, and GFP CFU per ×10 field were quantified at 6 days. (CPE denotes significant CPE at an adenovirus MOI of 10,000 particles/cell and was not quantified for GFP colonies.) Hirt DNAs from AV.GFP3ori (1,000 DNA particles/cell)-infected HeLa cells with or without coinfection with Ad.CMVlacZ (5,000 particles/cell) were used to transform E. coli. The total number of ampicillin-resistant bacterial CFU (D) and total number of head-to-tail circular intermediate CFU (E) are given for a representative experiment. More than 20 clones for each time point were evaluated by Southern blotting (see Fig. 2 for details). Zero hour control experiments were performed by mixing an amount of AV.GFP3ori virus equivalent to that used in experiments with mock-infected cellular lysates prior to Hirt DNA purification. (F) Abundance of head-to-tail circular intermediates as a percentage of total ampicillin-resistant bacterial CFU isolated from Hirt DNA.
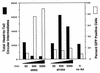
FIG. 4
Identification of adenovirus genes responsible for augmentation of AAV circular intermediate formation. HeLa cells were infected with AV.GFP3ori (1,000 DNA particles/cell) in the presence of _dl_802 (E2a deleted) and _dl_1004 (E4 deleted) adenovirus (at the indicated MOIs). The total number of head-to-tail circular intermediates from Hirt DNA and the level of augmentation of GFP transgene expression (as determined by FACS) were quantified at 24 h postinfection. Results are the average of duplicate experiments.
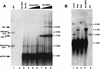
FIG. 5
Southern blot identification of circular and Rf rAAV genomes in Hirt DNA. To identify circular intermediates, one-half of the total Hirt DNA yield from a 35-mm-diameter plate of HeLa cells was loaded onto a 1% agarose gel and electrophoresed at 35 V for 14 h prior to Southern transfer. HeLa cell infections were carried out identically to that described in the legend to Fig. 4. GFP 32P-labeled probes were used for hybridization of Southern blots. The rAAV circular intermediate plasmid (p81) isolated from bacterial transformation was used as a marker for the migration of the supercoiled (closed circular) and relaxed (open circular or nicked) form molecules (indicated by open triangles in lane 1, panel A, and lane 5, panel B). A negative control of Hirt DNA isolated from uninfected parental HeLa cells was loaded in lane 2 of panel A. Also included in panel A are samples of HeLa cell Hirt DNA following rAAV infection alone (lane 3); coinfection with rAAV and Ad._dl_1004 at MOIs of 50, 500, and 5,000 (lanes 4, 5, and 6, respectively); and coinfection with rAAV and Ad._dl_801 at MOIs of 50, 500, and 5,000 (lanes 7, 8, and 9, respectively). The size of circular intermediates based on the migration of rescued bacterial plasmids is approximately 2.8 kb. Solid arrows mark replication form dimers (Rfd), replication form monomers (Rfm), and single-stranded AAV genomes (ssDNA). (B) Hirt DNA Southern blots from HeLa cells infected with rAAV alone prior to (lane 1) and following restriction digestion with _Ase_I (lane 2) and _Pst_I (lane 3). In addition to p81 as a marker for migration of rescued circular form genomes (lane 5), undigested Hirt DNA from cells coinfected with rAAV and Ad._dl_802 (MOI, 50) is also shown as a marker for replication form monomer (4.7 kb) linear-length dsDNA genomes.
FIG. 6
Model for independent mechanistic interactions of adenovirus with lytic- and latent-phase aspects of the AAV life cycle. The adenovirus E4 gene has been shown to augment the level of rAAV second-strand synthesis, giving rise to replication form dimers (Rfd) and monomers (Rfm). This augmentation leads to substantial increases in transgene expression from rAAV vectors and most closely mirrors lytic-phase replication of wtAAV as head-to-head and tail-to-tail concatemers. In contrast, E4 expression inhibits the formation of head-to-tail circular intermediates of AAV. Hence, it appears that increases in the amount of Rfd and Rfm dsDNA genomes do not increase the extent of circular intermediate formation. Such findings suggest that conversion of Rfm and Rfd to circular intermediates does not likely occur and implicate two mechanistically distinct pathways for their formation. In support of this hypothesis, adenovirus E2a gene expression does not enhance the formation of Rfm and Rfd genomes, but rather increases the abundance and/or stability of head-to-tail circular intermediates. Cm, monomer circular intermediate; Cd, dimer circular intermediate. Furthermore, in the absence of E4, E2a gene expression does not lead to augmentation of rAAV transgene expression. Given the similarities in genome structure of AAV circular intermediates to those of integrated proviruses, we hypothesize that circular form AAV genomes may represent preintegration complexes important in latent-phase persistence. In the presence of Rep, these circular intermediates may have a higher predisposition for integration into the host genome.
Similar articles
- Two independent molecular pathways for recombinant adeno-associated virus genome conversion occur after UV-C and E4orf6 augmentation of transduction.
Sanlioglu S, Duan D, Engelhardt JF. Sanlioglu S, et al. Hum Gene Ther. 1999 Mar 1;10(4):591-602. doi: 10.1089/10430349950018661. Hum Gene Ther. 1999. PMID: 10094202 - Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue.
Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher KJ, Engelhardt JF. Duan D, et al. J Virol. 1998 Nov;72(11):8568-77. doi: 10.1128/JVI.72.11.8568-8577.1998. J Virol. 1998. PMID: 9765395 Free PMC article. - The adenovirus E4 ORF6 and E1b 55 kDa proteins cooperate in a p53-independent manner to enhance transduction by recombinant adeno-associated virus vectors.
Trahair TN, Alexander IE, Rowe PB, Smythe JA. Trahair TN, et al. J Gen Virol. 2000 Dec;81(Pt 12):2983-2991. doi: 10.1099/0022-1317-81-12-2983. J Gen Virol. 2000. PMID: 11086129 - Integration of adeno-associated virus (AAV) and recombinant AAV vectors.
McCarty DM, Young SM Jr, Samulski RJ. McCarty DM, et al. Annu Rev Genet. 2004;38:819-45. doi: 10.1146/annurev.genet.37.110801.143717. Annu Rev Genet. 2004. PMID: 15568995 Review. - The cryptic life style of adeno-associated virus.
Berns KI, Linden RM. Berns KI, et al. Bioessays. 1995 Mar;17(3):237-45. doi: 10.1002/bies.950170310. Bioessays. 1995. PMID: 7748178 Review.
Cited by
- Hot topics in adeno-associated virus as a gene transfer vector.
Zhao N, Liu DP, Liang CC. Zhao N, et al. Mol Biotechnol. 2001 Nov;19(3):229-37. doi: 10.1385/MB:19:3:229. Mol Biotechnol. 2001. PMID: 11721619 Review. - Adeno-associated virus site-specific integration is mediated by proteins of the nonhomologous end-joining pathway.
Daya S, Cortez N, Berns KI. Daya S, et al. J Virol. 2009 Nov;83(22):11655-64. doi: 10.1128/JVI.01040-09. Epub 2009 Sep 16. J Virol. 2009. PMID: 19759155 Free PMC article. - Delivering Transgenic DNA Exceeding the Carrying Capacity of AAV Vectors.
Hirsch ML, Wolf SJ, Samulski RJ. Hirsch ML, et al. Methods Mol Biol. 2016;1382:21-39. doi: 10.1007/978-1-4939-3271-9_2. Methods Mol Biol. 2016. PMID: 26611576 Free PMC article. - Long-term inhibition of Hepatitis B virus gene expression by a primary microrna expressing ancestral adeno-associated viral vector.
Mnyandu NZ, Limani SW, Ely A, Wadee R, Arbuthnot P, Maepa MB. Mnyandu NZ, et al. Virol J. 2025 Feb 17;22(1):41. doi: 10.1186/s12985-025-02662-5. Virol J. 2025. PMID: 39962472 Free PMC article. Review.
References
- Bennett J, Duan D, Engelhardt J F, Maguire A M. Real-time, noninvasive in vivo assessment of adeno-associated virus mediated retinal transduction. Invest Ophthalmol Vis Sci. 1997;38:2857–2863. - PubMed
- Berns K I, Giraud C. Adeno-associated virus (AAV) vectors in gene therapy. Berlin, Germany: Springer; 1996.
- Conrad C K, Allen S S, Afione S A, Reynolds T C, Beck S E, Fee-Maki M, Barrazza-Ortiz X, Adams R, Askin F B, Carter B J, Guggino W B, Flotte T R. Safety of single-dose administration of an adeno-associated virus (AAV)-CFTR vector in the primate lung. Gene Ther. 1996;3:658–668. - PubMed
- Duan D, Fisher K J, Burda J F, Engelhardt J F. Structural and functional heterogeneity of integrated recombinant AAV genomes. Virus Res. 1997;48:41–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources