Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay - PubMed (original) (raw)
Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay
H Kimura et al. J Clin Microbiol. 1999 Jan.
Abstract
To measure the virus load in patients with symptomatic Epstein-Barr virus (EBV) infections, we used a real-time PCR assay to quantify the amount of EBV DNA in blood. The real-time PCR assay could detect from 2 to over 10(7) copies of EBV DNA with a wide linear range. We estimated the virus load in peripheral blood mononuclear cells (PBMNC) from patients with symptomatic EBV infections. The mean EBV-DNA copy number in the PBMNC was 10(3.7) copies/microg of DNA in patients with EBV-related lymphoproliferative disorders, 10(4.1) copies/microg of DNA in patients with chronic active EBV infections, and 10(2.2) copies/microg of DNA in patients with infectious mononucleosis. These numbers were significantly larger than those in either posttransplant patients or immunocompetent control patients without EBV-related diseases. In a patient with infectious mononucleosis, the virus load decreased as the symptoms resolved. The copy number of EBV DNA in PBMNC from symptomatic EBV infections was correlated with the EBV-positive cell number determined by the in situ hybridization assay (r = 0.842; P < 0.0001). These results indicate that the real-time PCR assay is useful for diagnosing symptomatic EBV infection and for monitoring the virus load.
Figures
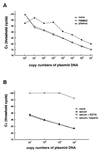
FIG. 1
(A) Standard curve for real-time PCR. Serially diluted pGEM-BALF5 plasmid was amplified with or without DNA extraction solutions from blood and analyzed in real time with a model 7700 Sequence Detector. The CT values were plotted against copy number to construct the standard curve. Explanation of symbols: none, plasmid with EBV insert; PBMNC, PBMNC from an EBV-seronegative patient plus plasmid with EBV insert; plasma, plasma from an EBV negative patient plus plasmid with EBV insert. (B) Effect of heparin on quantitation of plasmid DNA. Serially diluted plasmid controls were amplified with or without various DNA extraction solutions and analyzed using a model 7700 Sequence Detector. Explanation of symbols: none, plasmid with EBV insert; serum, serum from an EBV seronegative patient plus plasmid with EBV insert; serum + EDTA, serum and EDTA plus plasmid with EBV insert; serum + heparin, serum and heparin plus plasmid with EBV insert.
FIG. 2
Quantitation of EBV DNA by real-time PCR. DNA was extracted from PBMNC obtained from patients with symptomatic EBV infections or control patients without EBV-related diseases. Two hundred fifty nanograms of DNA was used for the real-time PCR assay, and the EBV DNA copy numbers per microgram of DNA are shown. Multiple samples for some patients were tested because repeated evaluations were needed. Bars show the means and standard deviations for each group. The dotted line shows the detection limit of the assay.
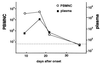
FIG. 3
Change in the virus load in a patient with IM. Sequential samples from a patient with IM were obtained, and both the PBMNC and plasma were analyzed by the real-time PCR assay. The EBV DNA copy numbers are shown per microgram of DNA. The EBV DNA copy numbers are shown per milliliter of plasma. The dotted line shows the detection limit for each sample.
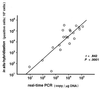
FIG. 4
Correlation of results of the real-time PCR and in situ hybridization assays. PBMNC were obtained from patients with symptomatic EBV diseases, and 20 samples were analyzed using both the real-time PCR and ISH assays. The copy numbers of EBV-DNA measured by the real-time PCR assay and EBV-positive cell numbers measured by the ISH assay were plotted, and the correlation coefficient (r) was calculated.
Similar articles
- Fluorescence in situ hybridization is superior for monitoring Epstein Barr viral load in infectious mononucleosis patients.
Cao P, Zhang M, Wang W, Dai Y, Sai B, Sun J, Wang L, Wang F, Li G, Xiang J. Cao P, et al. BMC Infect Dis. 2017 May 3;17(1):323. doi: 10.1186/s12879-017-2412-y. BMC Infect Dis. 2017. PMID: 28468603 Free PMC article. - Semiquantitative PCR analysis of Epstein-Barr virus DNA in clinical samples of patients with EBV-associated diseases.
Meerbach A, Gruhn B, Egerer R, Reischl U, Zintl F, Wutzler P. Meerbach A, et al. J Med Virol. 2001 Oct;65(2):348-57. doi: 10.1002/jmv.2040. J Med Virol. 2001. PMID: 11536243 - Real-time polymerase chain reaction for diagnosing infectious mononucleosis in pediatric patients: A systematic review and meta-analysis.
Jiang SY, Yang JW, Shao JB, Liao XL, Lu ZH, Jiang H. Jiang SY, et al. J Med Virol. 2016 May;88(5):871-6. doi: 10.1002/jmv.24402. Epub 2016 Jan 5. J Med Virol. 2016. PMID: 26455510 Review. - Measuring Epstein-Barr virus (EBV) load: the significance and application for each EBV-associated disease.
Kimura H, Ito Y, Suzuki R, Nishiyama Y. Kimura H, et al. Rev Med Virol. 2008 Sep-Oct;18(5):305-19. doi: 10.1002/rmv.582. Rev Med Virol. 2008. PMID: 18494041 Review.
Cited by
- Frequency of Epstein - Barr Virus in Patients Presenting with Acute Febrile Illness in Kenya.
Masakhwe C, Ochanda H, Nyakoe N, Ochiel D, Waitumbi J. Masakhwe C, et al. PLoS One. 2016 May 10;11(5):e0155308. doi: 10.1371/journal.pone.0155308. eCollection 2016. PLoS One. 2016. PMID: 27163791 Free PMC article. - Multiplex real-time PCR assay for simultaneous quantification of BK polyomavirus, JC polyomavirus, and adenovirus DNA.
Funahashi Y, Iwata S, Ito Y, Kojima S, Yoshikawa T, Hattori R, Gotoh M, Nishiyama Y, Kimura H. Funahashi Y, et al. J Clin Microbiol. 2010 Mar;48(3):825-30. doi: 10.1128/JCM.01699-09. Epub 2010 Jan 6. J Clin Microbiol. 2010. PMID: 20053854 Free PMC article. - Glucocorticoids activate Epstein Barr virus lytic replication through the upregulation of immediate early BZLF1 gene expression.
Yang EV, Webster Marketon JI, Chen M, Lo KW, Kim SJ, Glaser R. Yang EV, et al. Brain Behav Immun. 2010 Oct;24(7):1089-96. doi: 10.1016/j.bbi.2010.04.013. Epub 2010 May 11. Brain Behav Immun. 2010. PMID: 20466055 Free PMC article. - Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood.
Hakim H, Gibson C, Pan J, Srivastava K, Gu Z, Bankowski MJ, Hayden RT. Hakim H, et al. J Clin Microbiol. 2007 Jul;45(7):2151-5. doi: 10.1128/JCM.02308-06. Epub 2007 May 9. J Clin Microbiol. 2007. PMID: 17494720 Free PMC article. - Upregulation of Reg 1alpha and GW112 in the epithelium of inflamed colonic mucosa.
Shinozaki S, Nakamura T, Iimura M, Kato Y, Iizuka B, Kobayashi M, Hayashi N. Shinozaki S, et al. Gut. 2001 May;48(5):623-9. doi: 10.1136/gut.48.5.623. Gut. 2001. PMID: 11302958 Free PMC article.
References
- Baer R, Bankier A T, Biggin M D, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. - PubMed
- Bai X, Hosler G, Rogers B B, Dawson D B, Scheuermann R H. Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin Chem. 1997;43:1843–1849. - PubMed
- Beutler E, Gelbart T, Kuhl W. Interference of heparin with the polymerase chain reaction. BioTechniques. 1990;9:2–3. - PubMed
- Gan Y J, Sullivan J L, Sixbey J W. Detection of cell-free Epstein-Barr virus DNA in serum during acute infectious mononucleosis. J Infect Dis. 1994;170:436–439. - PubMed
- Gratama J W. Epstein-Barr virus infections in bone marrow transplantation recipients. In: Forman S J, Blume K G, Thomas E D, editors. Bone marrow transplantation. Boston, Mass: Blackwell Scientific Publications; 1994. pp. 429–442.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources