Fiber-type-specific transcription of the troponin I slow gene is regulated by multiple elements - PubMed (original) (raw)
Fiber-type-specific transcription of the troponin I slow gene is regulated by multiple elements
S Calvo et al. Mol Cell Biol. 1999 Jan.
Abstract
The regulatory elements that restrict transcription of genes encoding contractile proteins specifically to either slow- or fast-twitch skeletal muscles are unknown. As an initial step towards understanding the mechanisms that generate muscle diversity during development, we have identified a 128-bp troponin I slow upstream element (SURE) and a 144-bp troponin I fast intronic element (FIRE) that confer fiber type specificity in transgenic mice (M. Nakayama et al., Mol. Cell. Biol. 16:2408-2417, 1996). SURE and FIRE have maintained the spatial organization of four conserved motifs (3' to 5'): an E box, an AT-rich site (A/T2) that binds MEF-2, a CACC site, and a novel CAGG motif. Troponin I slow (TnIs) constructs harboring mutations in these motifs were analyzed in transiently and stably transfected Sol8 myocytes and in transgenic mice to assess their function. Mutations of the E-box, A/T2, and CAGG motifs completely abolish transcription from the TnI SURE. In contrast, mutation of the CACC motif had no significant effect in transfected myocytes or on the slow-specific transcription of the TnI SURE in transgenic mice. To assess the role of E boxes in fiber type specificity, a chimeric enhancer was constructed in which the E box of SURE was replaced with the E box from FIRE. This TnI E box chimera, which lacks the SURE NFAT site, confers essentially the same levels of transcription in transgenic mice as those conferred by wild-type SURE and is specifically expressed in slow-twitch muscles, indicating that the E box on its own cannot determine the fiber-type-specific expression of the TnI promoter. The importance of the 5' half of SURE, which bears little homology to the TnI FIRE, in muscle-specific expression was analyzed by deletion and linker scanning analyses. Removal of the 5' half of SURE (-846 to -811) results in the loss of expression in stably transfected but not in transiently expressing myocytes. Linker scanning mutations identified sequences in this region that are necessary for the function of SURE when integrated into chromatin. One of these sites (GTTAATCCG), which is highly homologous to a bicoid consensus site, binds to nuclear proteins from several mesodermal cells. These results show that multiple elements are involved in the muscle-specific activity of the TnIs promoter and that interactions between upstream and downstream regions of SURE are important for transcription in the context of native chromatin.
Figures
FIG. 1
Organization of the TnIs reporter constructs used in the studies. A schematic diagram of the 5′ flanking region of the TnIs gene is shown at the top. (A) TnIs reporter constructs used for cell transfections and the generation of transgenic mice. TnIs500 contains 479 bp of the TnIs promoter inserted upstream of either the CAT or the luciferase reporter coding sequence. TnIs500SURECAT and TnIs500SURELUC were constructed by the insertion of the 128-bp SURE fragment (−868 to −741) into the TnIs500 reporter. TnIs95LUC contains 95 bp of the basal TnIs promoter upstream of the luciferase reporter coding sequence, TnIs95SURE and TnIs95FIRE were generated by the insertion of SURE and the 144-bp FIRE fragment (+776 to +663 of quail TnIf intronic sequence), respectively, upstream of the TnIs95LUC basal promoter. The deletion constructs TnIs95SURE Δ−868/−778 and TnIs95 FIRE Δ+776/+703 were made by PCR amplification of fragments −778 to −741 of SURE and +703 to +633 of FIRE, respectively, and subsequent insertion into the _Sst_I-_Nhe_I-cleaved TnIs95LUC. (B) Schematic representation of the SURE-FIRE TnI E-box Chimera construct. Ebox Chimera was made by using the PCR ligation technique as described in Materials and Methods. The −868 to −759 sequence of SURE was joined to the +664 to +633 sequence of FIRE and inserted 5′ of the luciferase reporter. Open boxes, FIRE sequences; shaded boxes, SURE sequences.
FIG. 2
Mutational analysis of the conserved sites of SURE in stably transfected Sol8 myocytes. The transcriptional activities conferred by constructs harboring distinct mutations in each of the conserved motifs in TnI SURE were analyzed in stably transfected Sol8 myotubes. The levels of CAT reporter activity were measured in whole-cell extracts prepared from cells transfected with the A/T1, CAGG, CACC, A/T2, and E-box mutant constructs and compared to reporter levels conferred by the wild-type TnIs500SURE construct. The wild-type sequences from each of these motifs, and the mutations made therein, are shown at the bottom. For each construct, two independent DNA preparations were used to make the CaPO4 precipitates, and a total of six pools of stably transfected myotubes were analyzed per construct. The extracts (50 μg) were assayed for CAT activity at 37°C for 3 h under conditions of linear enzymatic activity. The values shown are means, and error bars indicate standard deviations.
FIG. 3
Analysis of the effects of mutations in the conserved sites of SURE in muscles of transgenic mice. CAT analyses were performed on extracts made from different muscles on transgenic mice. Independent transgenic mouse lines (indicated by number at the bottom of each bar graph) were generated with the wild-type TnIs500SURE fragment and fragments harboring mutations in the conserved A/T1, CAGG, CACC, and E-box motifs (see Fig. 2). The CAT assays were performed with 100 μg of extract at 37°C for 3 h under conditions of linear enzymatic activity. Since the values for CAT activities in the extracts of transgenic line 7575 of TnIs500SURE and 8764 of CACC were very high in relation to those of the other lines, these results are shown on a different scale.
FIG. 4
The E box from FIRE functionally substitutes for the SURE E box but does not change the fiber-type specificity of TnI SURE. Transgenic mouse lines were generated with a fragment from the TnI Ebox Chimera construct, in which the E box from SURE was swapped for the FIRE E box (Fig. 1). Luciferase activities assayed in extracts made from different hind-limb muscles of five TnI Ebox Chimera transgenic lines were normalized for protein concentration. The sequence of the TnI Ebox Chimera construct, at the boundary between SURE (shaded box) and FIRE (open box), is shown at the bottom; the E-box and A/T-2 motifs are shown in boldface.
FIG. 5
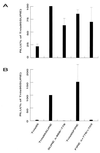
The 5′ halves of SURE and FIRE are important for transcriptional activity in stably transfected Sol8 myotubes. The transcriptional activities conferred by the constructs TnI95 (negative control), TnI95SURE (positive control), SURE Δ−868/−778 (5′ half deleted), and FIRE Δ+776/+703 (5′ half deleted) (see Fig. 1) were analyzed in transiently (A) and stably (B) transfected Sol8 myotubes. The luciferase reporter activities assayed in the extracts were normalized either to renilla activity expressed from an internal control plasmid (transient transfections) or for protein concentration (stable transfections). Values are means (n ≥ 6); error bars indicate standard deviations.
FIG. 6
EMSAs of nuclear protein binding to SURE upstream sequences. (A) Sequences between −840 and −829 of SURE interact with nuclear extracts from Sol8 myotubes. 32P-labeled double-stranded oligonucleotides with the wild-type sequence (SURE −850/−808WT) and GAATTC linker mutations (SURE −850/−808LS#3 to SURE −850/−808LS#8) are shown at the top (dashes indicate sequence identities between the wild-type and mutant oligonucleotides). The probe oligonucleotides were incubated on ice in the absence (−) or in the presence (+) of 5 μg of nuclear extracts from Sol8 myotubes. The positions of complexes I and II are indicated on the left. (B) The complexes that bind the −840 and −829 sequence are not restricted to muscle cells. Sequences of the SURE −842/−815, SURE −844/−827, and SURE −832/−815 oligonucleotides used in the EMSAs are shown at the top. Nuclear extracts (10 μg) from undifferentiated Sol8 myoblasts (MB), differentiated Sol8 myotubes (MT), HepG2 hepatoma cells, 3T3 fibroblasts, and cerebellar granule cells (CGN), as well as whole-tissue extracts (100 μg) from SOL and EDL hind-limb skeletal muscles were incubated with the 32P-labeled SURE −842/−815 oligonucleotide and analyzed as described above. For competition assays, a 100-fold molar excess of unlabeled SURE −842/−815, SURE −844/−827, or SURE −832/−815 was used. The SOL and EDL lanes were exposed to X-ray film for 72 h, and the other lanes were autoradiographed for 24 h.
FIG. 7
Linker scanning mutations through the 5′ end of TnI SURE. An _Eco_RI recognition sequence (GAATTC) was used to generate a series of linker scanning mutations in the upstream region of SURE (TnIsLS#3 to TnIsLS#9). Dashes indicate sequence identities between the wild-type and the mutant oligonucleotides. These constructs and wild-type TnIs500SURE were used to transfect Sol8 myocytes. Luciferase reporter activities were assayed in extracts made from Sol8 myotubes and normalized either for the renilla used as an internal control in the transient transfections (A) or for protein concentration in the stable transfections (B). Values are means (n ≥ 6); error bars indicate standard deviations.
Similar articles
- Transcriptional control of muscle plasticity: differential regulation of troponin I genes by electrical activity.
Calvo S, Stauffer J, Nakayama M, Buonanno A. Calvo S, et al. Dev Genet. 1996;19(2):169-81. doi: 10.1002/(SICI)1520-6408(1996)19:2<169::AID-DVG9>3.0.CO;2-7. Dev Genet. 1996. PMID: 8900050 - Molecular dissection of DNA sequences and factors involved in slow muscle-specific transcription.
Calvo S, Vullhorst D, Venepally P, Cheng J, Karavanova I, Buonanno A. Calvo S, et al. Mol Cell Biol. 2001 Dec;21(24):8490-503. doi: 10.1128/MCB.21.24.8490-8503.2001. Mol Cell Biol. 2001. PMID: 11713284 Free PMC article. - Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements.
Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Nakayama M, et al. Mol Cell Biol. 1996 May;16(5):2408-17. doi: 10.1128/MCB.16.5.2408. Mol Cell Biol. 1996. PMID: 8628309 Free PMC article. - The ups and downs of gene regulation by electrical activity in skeletal muscles.
Rana ZA, Gundersen K, Buonanno A. Rana ZA, et al. J Muscle Res Cell Motil. 2009 Dec;30(7-8):255-60. doi: 10.1007/s10974-010-9200-2. Epub 2010 Feb 5. J Muscle Res Cell Motil. 2009. PMID: 20135341 Free PMC article. Review. - Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex.
Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Lenka N, et al. Prog Nucleic Acid Res Mol Biol. 1998;61:309-44. doi: 10.1016/s0079-6603(08)60830-2. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9752724 Review.
Cited by
- Imaging transcription in vivo: distinct regulatory effects of fast and slow activity patterns on promoter elements from vertebrate troponin I isoform genes.
Rana ZA, Gundersen K, Buonanno A, Vullhorst D. Rana ZA, et al. J Physiol. 2005 Feb 1;562(Pt 3):815-28. doi: 10.1113/jphysiol.2004.075333. Epub 2004 Nov 4. J Physiol. 2005. PMID: 15528243 Free PMC article. - Activity-dependent repression of muscle genes by NFAT.
Rana ZA, Gundersen K, Buonanno A. Rana ZA, et al. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5921-6. doi: 10.1073/pnas.0801330105. Epub 2008 Apr 11. Proc Natl Acad Sci U S A. 2008. PMID: 18408153 Free PMC article. - Myocyte enhancer factor 2C function in skeletal muscle is required for normal growth and glucose metabolism in mice.
Anderson CM, Hu J, Barnes RM, Heidt AB, Cornelissen I, Black BL. Anderson CM, et al. Skelet Muscle. 2015 Feb 27;5:7. doi: 10.1186/s13395-015-0031-0. eCollection 2015. Skelet Muscle. 2015. PMID: 25789156 Free PMC article. - The role of muscle activation pattern and calcineurin in acetylcholinesterase regulation in rat skeletal muscles.
Pregelj P, Trinkaus M, Zupan D, Trontelj JJ, Sketelj J. Pregelj P, et al. J Neurosci. 2007 Jan 31;27(5):1106-13. doi: 10.1523/JNEUROSCI.4182-06.2007. J Neurosci. 2007. PMID: 17267565 Free PMC article. - Interaction between signalling pathways involved in skeletal muscle responses to endurance exercise.
Koulmann N, Bigard AX. Koulmann N, et al. Pflugers Arch. 2006 May;452(2):125-39. doi: 10.1007/s00424-005-0030-9. Epub 2006 Jan 18. Pflugers Arch. 2006. PMID: 16437222 Review.
References
- Apone S, Hauschka S D. Muscle gene E-box control elements. Evidence for quantitatively different transcriptional activities and the binding of distinct regulatory factors. J Biol Chem. 1995;270:21420–21427. - PubMed
- Ausubel F M. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. 2nd ed. Brooklyn, N.Y: Greene Publishing Associates; 1992.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials