Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins - PubMed (original) (raw)
Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins
G Hu et al. Mol Cell Biol. 1999 Jan.
Abstract
The Drosophila seven in absentia (sina) gene was initially discovered because its inactivation leads to R7 photoreceptor defects. Recent data indicate that Sina binds to the Sevenless pathway protein Phyllopod, and together they mediate degradation of Tramtrack, a transcriptional repressor of R7 cell fate. Independent studies have shown that Sina and its highly related mammalian homologues Siah-1 and Siah-2 bind to the DCC (deleted in colorectal cancer) protein and promote its proteolysis via the ubiquitin-proteasome pathway. To determine the roles of mammalian Siahs in proteolysis and their interactions with target proteins, we sought to define Siah-1 domains critical for regulation of DCC. Mutant Siah-1 proteins, harboring missense mutations in the carboxy (C)-terminal domain analogous to those present in Drosophila sina loss-of-function alleles, failed to promote DCC degradation. Point mutations and deletion of the amino (N)-terminal RING finger domain of Siah-1 abrogated its ability to promote DCC proteolysis. In the course of defining Siah-1 sequences required for DCC degradation, we found that Siah-1 is itself rapidly degraded via the proteasome pathway, and RING domain mutations stabilized the Siah-1 protein. Siah-1 was found to oligomerize with itself and other Sina and Siah proteins via C-terminal sequences. Finally, evidence that endogenous Siah-1 regulates DCC proteolysis in cells was obtained through studies of an apparent dominant negative mutant of Siah-1, as well as via an antisense approach. The data indicate that the Siah-1 N-terminal RING domain is required for its proteolysis function, while the C-terminal sequences regulate oligomerization and binding to target proteins, such as DCC.
Figures
FIG. 1
Schematic representation of Siah-1 and Sina proteins encoded by the expression vector constructs. For each protein, the locations of the N-terminal Flag (black box), HA (cross-hatched box), and c-Myc (hatched box) epitope tags are shown, as are the specific Siah or Sina sequences. Note that the full-length Siah-1 protein has 282 amino acids, and the full-length Drosophila Sina protein has 314 amino acids. The positions of the C-terminal missense substitutions are indicated, as are the sequences deleted in the various deletion mutants.
FIG. 2
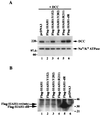
Regulation of DCC protein expression by wild-type and mutant Siah-1 proteins. (A) Wild-type Siah-1, but not certain mutated forms, promote DCC degradation. Western blot analysis of DCC expression in COS-1 cells cotransfected with pcDNA3 expression vectors containing the following cDNAs was done: DCC (lanes 1 to 6) and Flag epitope-tagged wild-type SIAH1 (lanes 2), Flag epitope-tagged and mutated SIAH1 cDNAs (lanes 3 to 6), or the empty pcDNA3 expression vector (lanes 1). Forty-eight hours after transfection, enhanced chemiluminescence (ECL)-Western blot analysis was carried out on the cell lysates, using the DCC extracellular domain monoclonal antibody G92-13. The membrane was then stripped, and Western blotting with a polyclonal antibody against Na+/K+ ATPase was performed to confirm equal loading of the lanes. The migration positions (in kilodaltons) of selected molecular mass markers are indicated to the left of the blots. (B) Western blot analysis of expression of wild-type (wt) and mutant (mts) forms of Siah-1. The cell lysates described in panel A were analyzed by ECL-Western blotting using the anti-Flag M2 monoclonal antibody. Extremely low levels of expression of the Flag-Siah-1 protein (lane 1) and three missense mutants (lanes 2 to 4) were detected only after long exposure to film. The strong signal in lane 5 represents the faster-migrating, RING-deleted form of Siah-1 (Flag-Siah-1-dR). The migration positions (in kilodaltons) of selected protein molecular mass markers are indicated to the right of the blot.
FIG. 3
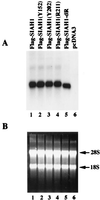
Reduced expression of wild-type Siah-1 protein and certain mutants is not due to reduced gene expression. (A) Northern blot analysis of the expression of SIAH1 transcripts following transfection of the SIAH1 cDNA constructs into COS-1 cells. The indicated SIAH1 expression constructs (lanes 1 to 5) or the empty vector control (lane 6) were transfected into COS-1 cells. Forth-eight hours after transfection, total RNA was extracted with Trizol reagent and Northern blotting was carried out. The blot was probed with a 32P-labeled SIAH1 cDNA fragment. Similar levels of wild-type and mutated SIAH1 transcripts were observed, in all lanes except the negative control (lane 6). (B) Ethidium bromide staining of the RNAs verifies equivalent loading. The 28S and 18S ribosomal bands are indicated.
FIG. 4
Regulation of Siah-1 protein expression by the proteasome pathway. (A) Increased expression of wild-type Siah-1 expression, but not the RING-deleted form, following treatment of cells with the proteasome inhibitor MG132. cDNAs encoding Flag-tagged wild-type Siah-1 and the RING-deleted form of Siah-1 (SIAH1-dR) were transfected into COS-1 cells, as indicated. Forty-eight hours after transfection, the cells were treated for 6 h with various concentrations of MG132, as indicated. Cell lysates were prepared, and ECL-Western blot analysis with the anti-Flag M2 antibody was performed. The blot was then stripped and reprobed with anti-actin polyclonal antibody C11 to verify the loading. The migration positions (in kilodaltons) of selected markers are shown to the left of the blots. (B) Expression of Siah-1 proteins with missense mutations is also increased following treatment of cells with MG132. Studies essentially identical to those in panel A were carried out. A no DNA “mock” negative-control transfection is shown in lane 9. The migration positions (in kilodaltons) of selected protein markers are shown to the left of the blots.
FIG. 5
Localized mutations in the RING domain abrogate the ability of Siah-1 to promote DCC degradation and result in stable expression of the mutant Siah-1 protein. ECL-Western blot studies of lysates from COS-1 cells harvested 48 h after cotransfection with an expression construct for DCC and a pcDNA3 expression construct containing no insert (vector) (lane 1), wild-type (wt) Siah-1 (lane 2), or a cDNA for a specific mutant Siah-1 protein with a single or double missense substitution in the RING domain (lanes 3 to 9) are shown. The various missense mutants of Siah-1 studied are indicated above the respective lane. DCC protein expression was detected with the G92-13 monoclonal antibody and expression of Siah-1 proteins was detected by Western blotting with an anti-Flag M2 monoclonal antibody reactive with the epitope tag at their N termini. The DCC and Flag Western blots were generated in parallel, and the Flag immunoblot was stripped and reprobed with an antibody against Na+/K+ ATPase to confirm equal loading of the lanes.
FIG. 6
In vitro and in vivo interactions of Sina and Siah proteins. (A) Wild-type and mutant Siah-1 proteins bind to DCC in vitro. In the upper panel, the products of coupled in vitro transcription and translation (IVTT) of wild-type and mutant SIAH1 cDNAs are shown. Essentially equivalent amounts of [35S]methinonine-labeled wild-type and mutated Siah-1 proteins were generated, and the proteins are Flag-Siah-1 (lane 1), Flag-Siah-1-dN (lane 2), Flag-Siah-1-dR (lane 3), Flag-Siah-1(Y152) (lane 4), Flag-Siah-1(Y202) (lane 5), Flag-Siah-1(R211) (lane 6), and a luciferase control (lane 7). In the lower panel, the in vitro binding of these proteins to a recombinant GST-DCC protein (lanes 1 to 8) is shown. In lane 8, the absence of binding of the Flag-Siah-1 protein to a control GST protein is shown. (B) In vitro binding of Siah-1 to the Sina protein. Wild-type and mutated Siah-1 proteins as well as a control luciferase protein were synthesized in vitro and are shown in the upper panel. The in vitro binding assay was carried out with a recombinant GST protein containing full Sina sequences (lower panel). (C) Siah-1 and Sina proteins form homo- and heterooligomers in cells. 293 cells were contransfected with expression constructs containing cDNAs for Flag-Siah-1-dR (Flag, amino acids [aa] 2 to 38 and 77 to 282), Flag-Siah-1-dN (Flag, aa 77 to 282), HA-Siah-1-dR (HA, aa 2 to 38 and 77 to 282), Myc-Sina (c-Myc-tagged full-length Sina), Myc-Sina-dC (Myc aa 2 to 199), or control expression vector (pcDNA3), as indicated. Forty-eight hours after transfection, cell lysates were prepared and a portion of lysates was used for immunoprecipitation with anti-Flag M2 monoclonal antibody. The cell lysates (Lysates) and immunoprecipitates (IPs) were electrophoresed, and Western blotting studies were carried out with anti-Flag M2, anti-HA 12CA5, and anti-c-Myc 9E10.2 monoclonal antibodies, respectively. Flag-tagged Siah-1 proteins coprecipitated with HA-tagged Siah-1-dR (lanes 7 and 8) as well as c-Myc-tagged Sina protein (lane 9). However, the Flag-tagged Siah-1 protein did not coprecipitate with a C-terminal truncated form of Sina (lane 10). The migration positions (in kilodaltons) of selected molecular mass markers are indicated to the left of the blots. IgG, immunoglobulin heavy chain.
FIG. 7
Oligomerization of Siah-1 proteins requires C-terminal sequences, and degradation function requires the RING finger domain. Regulation of HA-SIAH1-dR protein expression by SIAH1. COS-1 cells were cotransfected with constructs encoding an HA-tagged RING-deleted Siah-1 mutant (i.e., Siah-1-dR) (lanes 1 to 7) and Flag-tagged wild-type and mutant Siah-1 proteins (lanes 2 to 7, and indicated), or empty vector (lane 1). The expression of HA-Siah-1-dR was analyzed by Western blotting with anti-HA monoclonal antibody 12CA5. The same blot was then stripped and blotted with anti-actin polyclonal antibody C11 to verify equivalent loading in the lanes. The migration positions (in kilodaltons) of selected molecular mass markers are indicated to the left of the blots.
FIG. 8
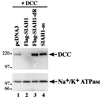
Endogenous Siah-1 functions to regulate DCC proteolysis in cells. Western blot studies were carried out on lysates of COS-1 cells cotransfected with a DCC expression construct and a pcDNA3 construct containing no cDNA insert (lane 1), a Flag-tagged wild-type SIAH-1 cDNA (lane 2), a Flag-tagged RING-deleted SIAH-1 cDNA (lane 3), or SIAH-1 cDNA in the antisense orientation (lane 4). Increased expression of DCC relative to the control lane was seen with the RING-deleted form of Siah-1 as well in the antisense studies. The blot was stripped and reprobed with an antibody against Na+/K+ ATPase to confirm equal loading. The migration positions (in kilodaltons) of selected molecular mass markers is indicated to the left of the blots.
FIG. 9
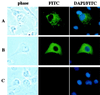
The RING domain regulates the localization of Siah-1 in cells. COS-1 cells were transiently transfected with expression plasmids encoding Flag-Siah-1 (A), Flag-Siah-1-dR (B), or an empty pcDNA3 vector as a control (C). Forty-eight hours after transfection, cells were incubated with Dulbecco’s modified Eagle medium containing 10% fetal bovine serum and 50 μM MG132 for 4 h at 37°C and then fixed. Cellular localization of the expressed proteins was detected by indirect immunofluorescence staining of the cells with anti-Flag M2 monoclonal antibody and a FITC-conjugated goat anti-mouse immunoglobulin secondary antibody. Cells were also stained with DAPI to visualize the nuclei.
References
- Blackwood M A, Weber B L. BRCA1 and BRCA2: from molecular genetics to clinical medicine. J Clin Oncol. 1998;16:1969–1977. - PubMed
- Borden K L B, Freemont P S. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. - PubMed
- Carthew R W, Rubin G M. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. - PubMed
- Chang H C, Solomon N M, Wassarman D A, Karim F D, Therrien M, Rubin G M, Wolff T. phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell. 1995;80:463–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases