Quorum-sensing acts at initiation of chromosomal replication in Escherichia coli - PubMed (original) (raw)
Quorum-sensing acts at initiation of chromosomal replication in Escherichia coli
H L Withers et al. Proc Natl Acad Sci U S A. 1998.
Abstract
Chromosomal replication in Escherichia coli was studied by flow cytometry and was found to be inhibited by an extracellular factor present in conditioned media collected during late exponential and early stationary phase, i.e., via a quorum-sensing mechanism. Our results suggest that the inhibitory activity of the extracellular factor is exerted during initiation of DNA replication rather than during elongation. Furthermore, we present evidence that this interaction may occur directly at each of the replication forks. Unlike other quorum-sensing systems described so far for Gram-negative bacteria, this inhibitory activity does not require transcription or translation to be effective. Implications of quorum-sensing regulation of DNA replication are discussed.
Figures
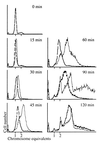
Figure 1
MG1655_dnaC2_ was synchronized for replication by incubation at nonpermissive temperature. At 0 min the culture was split into two and either no addition (solid line) was made or 1 ml of cCM per 10 ml of culture (dashed line) was added before the cultures were shifted to the permissive temperature. Samples were taken for flow cytometry at the times indicated.
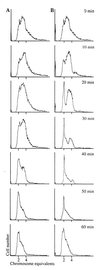
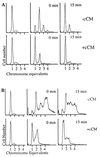
Figure 2
MG1655 was grown at 37°C to an OD600 of 0.1. The culture was split into two and either no addition (A) was made or 1 ml of cCM per 10 ml of culture (B) was added. Samples were taken for flow cytometry at the times indicated.
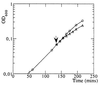
Figure 3
MG1655 was grown in M9 medium supplemented with glucose and Casamino acids at 37°C to the indicated OD (↓), whereupon the culture was split into two. Either 1 ml of sterile distilled water (○) or 1 ml of cCM (▵) per 10 ml of culture was added. Samples were taken at regular intervals throughout the experiment, and the OD was measured at 600 nm.
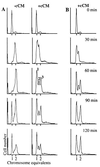
Figure 4
MG1655_dnaC2_ (A) and MG1655_dnaC2tus_ (B) were synchronized for replication by incubation at nonpermissive temperature. Both cultures were handled as follows. At 0 min the cultures were split into equal portions and either no addition was made or cCM was added to a final concentration of 2×. All cultures then were shifted to the permissive temperature for 10 min before being returned to the nonpermissive temperature for the duration of the experiment. Peaks 1, 2, and 3 referred to in the text are indicated on graphs where appropriate. Samples were taken for flow cytometry at the times indicated.
Figure 5
MG1655_dnaC2_ was synchronized by incubation at the nonpermissive temperature. At 0 min, the culture was divided into two equal parts. Either no addition (A) or cCM was added to a final concentration of 2× after the cultures were shifted to the permissive temperature (30°C) for 10 min and returned to the nonpermissive temperature (38°C) for a further 5-min incubation (B). Samples were taken for flow cytometry at the times indicated.
Figure 6
Analysis of transcriptional and translational requirements for inhibition of DNA replication by an extracellular factor. MG1655_dnaC2_ was synchronized at 38°C for 120 min. (A) After synchronization the culture was subdivided, and rifampin and cephalexin were added to both. At the same time cCM was added to one aliquot (Lower) to a final concentration of 2×; the other formed the control (Upper). In both cases, aliquots were incubated at 38°C for 0 or 15 min before being shifted to 30°C and incubated for a further 120 min before sampling for flow cytometry. (B) Same as for A except that chloramphenicol was added instead of rifampin.
Similar articles
- Obg/CtgA, a signaling protein that controls replication, translation, and morphological development?
Michel B. Michel B. Dev Cell. 2005 Mar;8(3):300-1. doi: 10.1016/j.devcel.2005.02.002. Dev Cell. 2005. PMID: 15737924 - Different effects of mioC transcription on initiation of chromosomal and minichromosomal replication in Escherichia coli.
Løbner-Olesen A, Boye E. Løbner-Olesen A, et al. Nucleic Acids Res. 1992 Jun 25;20(12):3029-36. doi: 10.1093/nar/20.12.3029. Nucleic Acids Res. 1992. PMID: 1620598 Free PMC article. - Host controlled plasmid replication: Escherichia coli minichromosomes.
Dasgupta S, Løbner-Olesen A. Dasgupta S, et al. Plasmid. 2004 Nov;52(3):151-68. doi: 10.1016/j.plasmid.2004.08.001. Plasmid. 2004. PMID: 15518873 Review. - Overexpression of the cgtA (yhbZ, obgE) gene, coding for an essential GTP-binding protein, impairs the regulation of chromosomal functions in Escherichia coli.
Dutkiewicz R, Slomińska M, Wegrzyn G, Czyz A. Dutkiewicz R, et al. Curr Microbiol. 2002 Dec;45(6):440-5. doi: 10.1007/s00284-002-3713-x. Curr Microbiol. 2002. PMID: 12402086 - [Novel function of DNA polymerase in negative regulation of initiation of chromosomal replication].
Katayama T. Katayama T. Tanpakushitsu Kakusan Koso. 1999 Jun;44(7):845-53. Tanpakushitsu Kakusan Koso. 1999. PMID: 10380576 Review. Japanese. No abstract available.
Cited by
- Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression.
Weeks JN, Galindo CL, Drake KL, Adams GL, Garner HR, Ficht TA. Weeks JN, et al. BMC Microbiol. 2010 Jun 8;10:167. doi: 10.1186/1471-2180-10-167. BMC Microbiol. 2010. PMID: 20529360 Free PMC article. - Small molecule signaling, regulation, and potential applications in cellular therapeutics.
McNerney MP, Styczynski MP. McNerney MP, et al. Wiley Interdiscip Rev Syst Biol Med. 2018 Mar;10(2):10.1002/wsbm.1405. doi: 10.1002/wsbm.1405. Epub 2017 Sep 28. Wiley Interdiscip Rev Syst Biol Med. 2018. PMID: 28960879 Free PMC article. Review. - Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence.
Day WA Jr, Maurelli AT. Day WA Jr, et al. Infect Immun. 2001 Jan;69(1):15-23. doi: 10.1128/IAI.69.1.15-23.2001. Infect Immun. 2001. PMID: 11119484 Free PMC article. - Inoculum size influences bacterial cross contamination between surfaces.
Montville R, Schaffner DW. Montville R, et al. Appl Environ Microbiol. 2003 Dec;69(12):7188-93. doi: 10.1128/AEM.69.12.7188-7193.2003. Appl Environ Microbiol. 2003. PMID: 14660365 Free PMC article. - Evolutionary genome engineering using a restriction-modification system.
Asakura Y, Kojima H, Kobayashi I. Asakura Y, et al. Nucleic Acids Res. 2011 Nov 1;39(20):9034-46. doi: 10.1093/nar/gkr585. Epub 2011 Jul 23. Nucleic Acids Res. 2011. PMID: 21785135 Free PMC article.
References
- Fuqua C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. - PubMed
- Swift S, Throup J P, Williams P, Salmond G P C, Stewart G S A B. Trends Biochem Sci. 1996;21:214–219. - PubMed
- Engebrecht J, Nealson K H, Silverman M. Cell. 1983;32:773–781. - PubMed
- Bainton N J, Bycroft B W, Chabra S R, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P. Gene. 1992;116:87–91. - PubMed
- Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources