Repair of oxidized bases in the extremely radiation-resistant bacterium Deinococcus radiodurans - PubMed (original) (raw)
Repair of oxidized bases in the extremely radiation-resistant bacterium Deinococcus radiodurans
C Bauche et al. J Bacteriol. 1999 Jan.
Abstract
Deinococcus radiodurans is able to resist and survive extreme DNA damage induced by ionizing radiation and many other DNA-damaging agents. It is believed that it possesses highly efficient DNA repair mechanisms. To characterize the repair pathway of oxidized purines in this bacteria, we have purified, from crude extracts, proteins that recognize these oxidized bases. We report here that D. radiodurans possesses two proteins excising the oxidized purines (formamidopyrimidine and 8-oxoguanine) by a DNA glycosylase-a purinic/apyrimidine lyase mechanism. Moreover, one of those proteins is endowed with a thymine glycol DNA glycosylase activity. One of these proteins could be the homolog of the Escherichia coli Fpg enzyme, which confirms the existence of a base excision repair system in this bacteria.
Figures
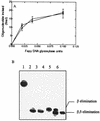
FIG. 1
Cleavage of the 8-oxoG/C duplex by the following: lane 1, no enzyme, lane 2, 5 ng of E. coli Fpg protein; lanes 3, 4, and 5, 0.2, 0.4, and 1 U of Fapy DNA glycosylase (fraction V); lanes 6, 7, and 8, 0.2, 0.4, and 1 U of Fapy DNA glycosylase (fraction IV). The duplex (100 fmol/reaction) was incubated with the enzyme at 32°C for 10 min.
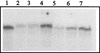
FIG. 2
Immunoblotting of fractions V and VI protein with anti-Fpg (A) and anti-Nth (B) antibodies. Lane 1, E. coli Fpg protein (100 ng); lane 2, E. coli Nth protein (100 ng); lane 3, fraction V (0.5 U of Fapy-DNA glycosylase); lane 4, fraction VI (0.5 U of Fapy-DNA glycosylase).
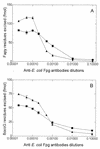
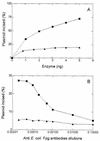
FIG. 3
D. radiodurans Fapy and 8-oxoG DNA glycosylase activities in the presence of various concentrations of anti-Fpg antibodies. Fraction V (0.04 U of Fapy DNA glycosylase; ■) and fraction VI (0.04 U of Fapy DNA glycosylase; ▴) were incubated for 10 min at 32°C with 700 fmol of [3H]Fapy–poly(dG-dC) or 100 fmol of 8-oxoG/C-containing oligonucleotide (34-mer) (B).
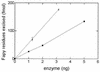
FIG. 4
Fapy-DNA glycosylase activities of fraction V (■) and fraction VI (▴). Increasing amounts of each enzyme were incubated in the presence of [3H]Fapy–poly(dG-dC) at 32°C for 10 min, and the [3H]Fapy residues released were measured (for details, see Materials and Methods).
FIG. 5
Cleavage of DNA duplexes containing a single 8-oxoG mismatched with one of the four bases C (■), T (▾), G (▴), and A (•). The various duplexes (100 fmol) were incubated with increasing amounts of proteins standardized as Fapy-DNA glycosylase activity of fraction V (A) and fraction VI (B) for 10 min at 32°C.
FIG. 6
NaBH4 trapping of 8-oxoG-containing DNA (200 fmol) treated with different amounts of proteins standardized as Fapy-DNA glycosylase activity from fraction V (■) and fraction VI (▴). The reactions were performed at 32°C for 30 min (for details, see Materials and Methods).
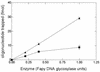
FIG. 7
Cleavage of an oligonucleotide containing a unique abasic site. Oligonucleotide (100 fmol) was incubated for 10 min at 32°C with enzyme. The products of the reaction were separated by SDS-PAGE, and the results were quantified with PhosphorImager. (A) Increasing amounts of fraction V (■) and fraction VI (▴). (B) Lane 1, duplex oligonucleotide containing an AP site. lane 2, like lane 1 but incubated with 0.2 M NaOH before analysis; lane 3, like lane 1 but incubated with 10 ng of E. coli Fpg protein; lane 4, like lane 1 but incubated with 10 ng of E. coli Nth protein; lane 5, like lane I but incubated with 10 ng of fraction V enzyme; lane 6, like lane 1 but incubated with 10 ng of fraction VI enzyme.
FIG. 8
NaBH4 trapping of AP site-containing DNA (100 fmol) with 50 ng of E. coli Fpg protein (lane 1), 10, 20 50 ng of fraction V enzyme (lanes 2, 3, and 4), and 10, 20, and 50 ng of fraction VI enzyme (lanes 5, 6, and 7) for 10 min at 32°C.
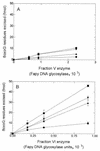
FIG. 9
Incision of OsO4-treated DNA by the D. radiodurans enzymes. (A) 500 ng of OsO4-treated DNA was incubated at 32°C for 10 min with increasing amounts of fraction V (■) and fraction VI (▴) enzymes. The products of the reaction were separated on an agarose gel and quantified by microdensitometric analysis (for details, see Materials and Methods). (B) Thymine glycol DNA glycosylase activities of fraction V (■) and fraction VI (▴) enzymes in the presence of different dilutions of anti E. coli Fpg antibodies.
Similar articles
- Substrate specificity of Deinococcus radiodurans Fpg protein.
Sentürker S, Bauche C, Laval J, Dizdaroglu M. Sentürker S, et al. Biochemistry. 1999 Jul 20;38(29):9435-9. doi: 10.1021/bi990680m. Biochemistry. 1999. PMID: 10413519 - Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli.
Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Hazra TK, et al. Nucleic Acids Res. 2001 May 1;29(9):1967-74. doi: 10.1093/nar/29.9.1967. Nucleic Acids Res. 2001. PMID: 11328881 Free PMC article. - Enzymatic properties of Escherichia coli and human 7,8-dihydro-8-oxoguanine DNA glycosylases.
Asagoshi K, Yamada T, Terato H, Ohyama Y, Ide H. Asagoshi K, et al. Nucleic Acids Symp Ser. 2000;(44):11-2. doi: 10.1093/nass/44.1.11. Nucleic Acids Symp Ser. 2000. PMID: 12903244 - Properties and biological functions of the NTH and FPG proteins of Escherichia coli: two DNA glycosylases that repair oxidative damage in DNA.
Boiteux S. Boiteux S. J Photochem Photobiol B. 1993 Jul;19(2):87-96. doi: 10.1016/1011-1344(93)87101-r. J Photochem Photobiol B. 1993. PMID: 8377077 Review. - Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions.
Hazra TK, Hill JW, Izumi T, Mitra S. Hazra TK, et al. Prog Nucleic Acid Res Mol Biol. 2001;68:193-205. doi: 10.1016/s0079-6603(01)68100-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554297 Review.
Cited by
- Oxidative stress resistance in Deinococcus radiodurans.
Slade D, Radman M. Slade D, et al. Microbiol Mol Biol Rev. 2011 Mar;75(1):133-91. doi: 10.1128/MMBR.00015-10. Microbiol Mol Biol Rev. 2011. PMID: 21372322 Free PMC article. Review. - Molecular cloning and functional analysis of the MutY homolog of Deinococcus radiodurans.
Li X, Lu AL. Li X, et al. J Bacteriol. 2001 Nov;183(21):6151-8. doi: 10.1128/JB.183.21.6151-6158.2001. J Bacteriol. 2001. PMID: 11591657 Free PMC article. - Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA.
Matsumoto Y, Zhang QM, Takao M, Yasui A, Yonei S. Matsumoto Y, et al. Nucleic Acids Res. 2001 May 1;29(9):1975-81. doi: 10.1093/nar/29.9.1975. Nucleic Acids Res. 2001. PMID: 11328882 Free PMC article. - Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags.
Lipton MS, Pasa-Tolic' L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic' N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Lipton MS, et al. Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11049-54. doi: 10.1073/pnas.172170199. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177431 Free PMC article. - Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics.
Dizdaroglu M, Coskun E, Jaruga P. Dizdaroglu M, et al. Mutat Res Rev Mutat Res. 2017 Jan-Mar;771:99-127. doi: 10.1016/j.mrrev.2017.02.001. Epub 2017 Feb 16. Mutat Res Rev Mutat Res. 2017. PMID: 28342455 Free PMC article. Review.
References
- Anderson A W, Norden H C, Cain R F, Parrish G, Duggan D. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 1956;10:575–578.
- Asahara H, Wistort P M, Bank J F, Bakerian R H, Cunningham R P. Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry. 1989;28:4444–4449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources