Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies - PubMed (original) (raw)
Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies
H J Chial et al. J Cell Biol. 1998.
Abstract
We report a novel connection between nuclear pore complexes (NPCs) and spindle pole bodies (SPBs) revealed by our studies of the Saccharomyces cerevisiae NDC1 gene. Although both NPCs and SPBs are embedded in the nuclear envelope (NE) in yeast, their known functions are quite distinct. Previous work demonstrated that NDC1 function is required for proper SPB duplication (Winey, M., M.A. Hoyt, C. Chan, L. Goetsch, D. Botstein, and B. Byers. 1993. J. Cell Biol. 122:743-751). Here, we show that Ndc1p is a membrane protein of the NE that localizes to both NPCs and SPBs. Indirect immunofluorescence microscopy shows that Ndc1p displays punctate, nuclear peripheral localization that colocalizes with a known NPC component, Nup49p. Additionally, distinct spots of Ndc1p localization colocalize with a known SPB component, Spc42p. Immunoelectron microscopy shows that Ndc1p localizes to the regions of NPCs and SPBs that interact with the NE. The NPCs in ndc1-1 mutant cells appear to function normally at the nonpermissive temperature. Finally, we have found that a deletion of POM152, which encodes an abundant but nonessential nucleoporin, suppresses the SPB duplication defect associated with a mutation in the NDC1 gene. We show that Ndc1p is a shared component of NPCs and SPBs and propose a shared function in the assembly of these organelles into the NE.
Figures
Figure 1
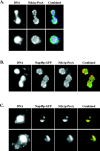
Ndc1p-ProA localizes to NPCs in whole cells. (A) Indirect IF deconvolution microscopy (see Materials and Methods) of yeast strain ProA5-4d/4a (Table I). Ndc1p-ProA was indirectly labeled using an FITC-conjugated secondary antibody, and DNA was visualized using 4′,6-diamidino-2-phenylindole. (B) Indirect IF deconvolution microscopy of yeast strain HC26-15a/1a (Table I) containing Ndc1p-ProA and Nup49p-GFP (Bucci and Wente, 1997). Ndc1p-ProA was indirectly labeled using a Texas red–conjugated secondary antibody, and Nup49p-GFP autofluorescence was detected using FITC filters. The combined image shows both Texas red (red) and GFP (green) signal (overlap = yellow). (C) Indirect IF deconvolution microscopy of Ndc1p-ProA localization and Nup49p-GFP localization in the presence of the nup133 null allele, which causes NPCs to cluster (HC27-16d/16b; Table I) (Doye et al., 1994; Pemberton et al., 1995). Ndc1p-ProA and Nup49p-GFP were detected as in B. The combined image shows both Texas red (red) and GFP (green) signal (overlap = yellow).
Figure 2
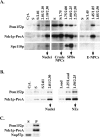
Biochemical analysis of Ndc1p-ProA. (A) Biochemical fractionation was carried out using a yeast strain that contains Ndc1p-ProA (ProA5-4d; Table I) (Rout and Kilmartin, 1990; Rout and Kilmartin, 1991; Rout and Blobel, 1993). Fractions containing nuclei, crude NPCs, SPBs, and enriched NPCs (E-NPCs) are labeled. Individual fractions were analyzed by SDS-PAGE and immunoblotted using antibodies to detect Pom152p, Ndc1p-ProA, or Spc110p. Pom152p is a NPC component (Wozniak et al., 1994), and Spc110p is a SPB component (Rout and Kilmartin, 1990; Kilmartin et al., 1993). Cell equivalents loaded were 1n for the left panels and 10n for the middle and right panels. (B) NEs were isolated from the Ndc1p-ProA yeast strain (ProA5-4d; Table I) using a previously described technique (Strambio-de-Castillia et al., 1995). Cell equivalents loaded were 1n for the left panels and 3n for the right panels. (C) Samples from the NE fraction were extracted with carbonate followed by centrifugation to determine whether Ndc1p-ProA was found in the supernatant (S) or in the membrane-containing pellet (P) (see Materials and Methods). Samples were analyzed by SDS-PAGE and immunoblotted using antibodies to detect Ndc1p-ProA, Pom152p, or Nup57p.
Figure 3
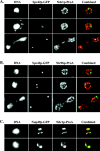
Ndc1p-ProA localizes to SPBs in whole cells. (A) Indirect IF deconvolution microscopy of cells (HC22-2b/1c; Table I) containing Ndc1p-ProA and Spc42p-GFP. Ndc1p-ProA was detected using a Texas red–conjugated secondary antibody, and Spc42p-GFP was detected by means of GFP autofluorescence using FITC filters (see Materials and Methods). The combined image shows both Texas red (red) and FITC (green) signal (overlap = yellow). (B) Indirect IF deconvolution microscopy of cells containing Nic96p-ProA and Spc42p-GFP (HC23-11d/16a; Table I) serves as a negative control for A. Samples were prepared as in A. The combined image shows both Texas red (red) and FITC signal (green). (C) Indirect IF deconvolution microscopy to examine the colocalization of Ndc1p-ProA with Nup49p-GFP in a nup133 null strain that exhibits NPC clustering (HC27-16d/16b; Table I). Ndc1p-ProA was detected as in A, and Nup49p-GFP was detected using FITC filters. The combined image shows both Texas red (red) and FITC (green) signals (overlap = yellow). Shown here are examples of cells that contain either one or two extra spots of Ndc1p-ProA staining that do not colocalize with Nup49p-GFP signal.
Figure 4
Ndc1p-GFP localizes to the membrane interacting regions of NPCs and SPBs. Ndc1p-GFP localization was examined using a diploid strain that is homozygous for the ndc1 null allele and is transformed with a centromeric plasmid containing NDC1-GFP [HC14-10c (1198)/HC29-6b; Table I]. Ndc1p-GFP localization was observed in living cells using deconvolution fluorescence microscopy (A). Additionally, this localization was examined using immunoelectron microscopy of whole cells (see Materials and Methods). Immunogold labeling, corresponding to Ndc1p-GFP, was detected at the periphery of NPCs (B) and at the edges of the central plaque region of SPBs (C); these are the regions of NPCs and SPBs that interact with the NE.
Figure 5
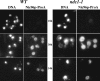
ndc1-1 strains do not exhibit defects in NPC assembly. An assay to examine NPC assembly was carried out using either wild-type (WT) or ndc1-1 cells (HC10-42a/42d and HC10-42b/42c, respectively; Table I) transformed with a plasmid containing NIC96-ProA expressed under the control of a galactose-inducible promoter (pGAL-NIC96-ProA). Asynchronous cultures were shifted to 13.5°C for 18 h (18h); at this point, the ndc1-1 culture contained 80% large budded cells. NIC96-ProA expression was induced for 12 h (30h), followed by a glucose chase for 4 h to determine whether Nic96p-ProA was properly assembled into NPCs (34h). Nic96p-ProA localization was visualized using indirect IF microscopy with an FITC-conjugated secondary antibody (see Materials and Methods).
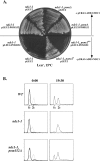
Figure 6
Deletion of POM152 suppresses the ndc1-1 SPB duplication defect in the first cell cycle. (A) ndc1-1 or ndc1-1, pom152 null double mutant strains [HC5-31c and HC5-31c(1166), respectively; Table I] containing pURA3-NDC1 (pALR10-NDC1, see Materials and Methods) or lacking pURA3-NDC1 were transformed with either pRS315 (pLEU2) or pPM1-HA (pLEU2-POM152) and were streaked to a 15°C Leu− plate. All strains containing pURA3-NDC1 were able to grow at the nonpermissive temperature. The ndc1-1 strain lacking pURA3-NDC1 failed to grow. However, the ndc1-1, pom152 null double mutant strain grew at 15°C when it contained the pLEU2 vector (*). When a plasmid-borne copy of POM152 (pLEU2-POM152) was reintroduced, the double mutant strain exhibited the ndc1-1 cold-sensitive phenotype (**). (B) DNA content of wild-type (WT), ndc1-1, and ndc1-1, pom152 null double mutant (ndc1-1, pom152Δ) strains [HC14-10c, HC5-31c, and HC5-31c(1166), respectively; Table I]. Asynchronous cultures (0:00, dashed lines) were arrested in G1 (1c DNA content; G2 and M cells have 2c DNA content) by treatment with alpha-factor at the permissive temperature (0:00, bold line) (see Materials and Methods). Cells were released from this arrest into media at the nonpermissive temperature for ndc1-1 for 19.5 h (19:30).
Similar articles
- A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly.
Lau CK, Giddings TH Jr, Winey M. Lau CK, et al. Eukaryot Cell. 2004 Apr;3(2):447-58. doi: 10.1128/EC.3.2.447-458.2004. Eukaryot Cell. 2004. PMID: 15075274 Free PMC article. - The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p.
Araki Y, Lau CK, Maekawa H, Jaspersen SL, Giddings TH Jr, Schiebel E, Winey M. Araki Y, et al. Mol Biol Cell. 2006 Apr;17(4):1959-70. doi: 10.1091/mbc.e05-07-0668. Epub 2006 Jan 25. Mol Biol Cell. 2006. PMID: 16436507 Free PMC article. - Integrity and function of the Saccharomyces cerevisiae spindle pole body depends on connections between the membrane proteins Ndc1, Rtn1, and Yop1.
Casey AK, Dawson TR, Chen J, Friederichs JM, Jaspersen SL, Wente SR. Casey AK, et al. Genetics. 2012 Oct;192(2):441-55. doi: 10.1534/genetics.112.141465. Epub 2012 Jul 13. Genetics. 2012. PMID: 22798490 Free PMC article. - Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.
Jaspersen SL, Ghosh S. Jaspersen SL, et al. Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review. - Big Lessons from Little Yeast: Budding and Fission Yeast Centrosome Structure, Duplication, and Function.
Cavanaugh AM, Jaspersen SL. Cavanaugh AM, et al. Annu Rev Genet. 2017 Nov 27;51:361-383. doi: 10.1146/annurev-genet-120116-024733. Epub 2017 Sep 15. Annu Rev Genet. 2017. PMID: 28934593 Review.
Cited by
- Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase.
Jones MH, Bachant JB, Castillo AR, Giddings TH Jr, Winey M. Jones MH, et al. Mol Biol Cell. 1999 Jul;10(7):2377-91. doi: 10.1091/mbc.10.7.2377. Mol Biol Cell. 1999. PMID: 10397771 Free PMC article. - A ternary membrane protein complex anchors the spindle pole body in the nuclear envelope in budding yeast.
Kupke T, Malsam J, Schiebel E. Kupke T, et al. J Biol Chem. 2017 May 19;292(20):8447-8458. doi: 10.1074/jbc.M117.780601. Epub 2017 Mar 29. J Biol Chem. 2017. PMID: 28356353 Free PMC article. - Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae.
Witkin KL, Friederichs JM, Cohen-Fix O, Jaspersen SL. Witkin KL, et al. Genetics. 2010 Nov;186(3):867-83. doi: 10.1534/genetics.110.119149. Epub 2010 Aug 16. Genetics. 2010. PMID: 20713690 Free PMC article. - The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p.
Castillo AR, Meehl JB, Morgan G, Schutz-Geschwender A, Winey M. Castillo AR, et al. J Cell Biol. 2002 Feb 4;156(3):453-65. doi: 10.1083/jcb.200111025. Epub 2002 Feb 4. J Cell Biol. 2002. PMID: 11827982 Free PMC article. - Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation.
Kerscher O, Hieter P, Winey M, Basrai MA. Kerscher O, et al. Genetics. 2001 Apr;157(4):1543-53. doi: 10.1093/genetics/157.4.1543. Genetics. 2001. PMID: 11290711 Free PMC article.
References
- Ausubel, J.D., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1994. Current Protocols in Molecular Biology. John Wiley and Sons, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases