Complete inhibition of Cdk/cyclin by one molecule of p21(Cip1) - PubMed (original) (raw)
Complete inhibition of Cdk/cyclin by one molecule of p21(Cip1)
L Hengst et al. Genes Dev. 1998.
Abstract
Cell-cycle phase transitions are controlled by cyclin-dependent kinases (Cdks). Key to the regulation of these kinase activities are Cdk inhibitors, proteins that are induced in response to various antiproliferative signals but that can also oscillate during cell-cycle progression, leading to Cdk inactivation. A current dogma is that kinase complexes containing the prototype Cdk inhibitor p21 transit between active and inactive states, in that Cdk complexes associated with one p21 molecule remain active until they associate with additional p21 molecules. However, using a number of different techniques including analytical ultracentrifugation of purified p21/cyclin A/Cdk2 complexes we demonstrate unambiguously that a single p21 molecule is sufficient for kinase inhibition and that p21-saturated complexes contain only one stably bound inhibitor molecule. Even phosphorylated forms of p21 remain efficient inhibitors of Cdk activities. Therefore the level of Cdk inactivation by p21 is determined by the fraction of kinase complexed with the inhibitor and not by the stoichiometry of inhibitor bound to the kinase or the phosphorylation state of the Cdk inhibitor.
Figures
Figure 1
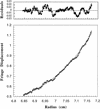
Free p21 is a monomer. The buoyant molecular mass of recombinant p21 was determined by sedimentation equilibrium analytical ultracentrifugation and is 18,490 ± 772 daltons. A scan after 30 hr is shown. (Bottom) The raw concentration data determined using interference optics and the solid line drawn through the data points was obtained by fitting the fringe displacement vs. radial position to a single species model of equation 1 (see Materials and Methods). (Top) The residual difference between the experimental data and the fitted data for each point shown.
Figure 2
Inhibition of cyclin A/Cdk2 by immobilized p21. (lanes 1–3) or anti-cyclin A (lane 4) antibodies were bound to protein A–Sepharose beads. These beads were incubated with varying amounts of purified p21: (lane 1) 0.05 μg; (lane 2) 0.2 μg; (lane 3) 0.2 μg; (lane 4) 0 μg. All beads were washed and incubated with ∼ 0.2 μg cyclin A/Cdk2. Nonbound kinase was removed from the beads and the sample of lane 3 was incubated subsequently with additional 0.4 μg of p21. The nonbound p21 was removed and the kinase activities associated with all samples were determined using [γ-32P]ATP and histone H1 as substrates. (Left) The amount of precipitated p21 protein (anti-p21) and Cdk2 (anti-PSTAIRE) was determined in Western blots using monoclonal antibodies. The level of 32P incorporation in the histone H1 bands (histone H1) was determined after SDS-PAGE by autoradiography of the dried gel. The amount of 32P incorporation in the histone H1 bands shown in this autoradiograph was measured using a PhosphorImager. The quantities of precipitated cyclin A and Cdk2 were determined by densitometric scanning of anti-PSTAIRE and anti-cyclin A Western blots and the histone H1 kinase activity of the samples is shown after normalization for the amount of precipitated kinase protein (right).
Figure 3
Size-exclusion chromatography of cyclin A/Cdk2 (top) and p21-saturated complexes of cyclin A/Cdk2 (bottom). Proteins were resolved on a Superdex 200 FPLC column. Aliquots of each fraction were analyzed by SDS-PAGE and stained using Coomassie blue. The migration of size standards are shown at the left on the gel, and the elution behavior, the Stokes radius, and the apparent molecular mass of marker proteins separated on this column are shown above.
Figure 4
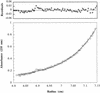
Determination of the molecular mass of purified cyclin A/Cdk2 by sedimentation equilibrium centrifugation. The apparent molecular mass of the cyclin A/Cdk2 kinase complex was determined as 86 ± 2 daltons. (Bottom) The raw concentration data determined by measuring the absorbance at 235 nm (cyclin A/Cdk2). The solid line drawn through the data points was obtained by fitting the fringe displacement vs. radial position to a single-species model. (Top) The residual differences between the experimental data and the fitted data for each point. The purified proteins used for analytical ultracentrifugation are shown in Fig. 6. Sedimentation equilibrium analysis was performed at 4°C and a rotor speed of 10,000 rpm.
Figure 5
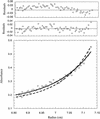
Determination of the molecular mass of purified p21/cyclin A/Cdk2 complexes. The apparent molecular mass of cyclin A/Cdk2 complex increases from 86 to 105 kD after saturation of the complex with p21. (Bottom) The raw concentration data determined by measuring the absorbance at 280 nm. The solid line drawn through the data points was obtained by fitting the fringe displacement vs. radial position to a single-species model. The dashed line represents the predicted behavior of a 1:1:2 (cyclin A:Cdk2:p21) complex using the same single-species model. (Top panels) The residual differences between the experimental data and the fitted data for each point, with those corresponding to the 1:1:2 model placed above those corresponding to the 1:1:1 model. The purified proteins used for analytical ultracentrifugation are shown in Fig.6. Sedimentation equilibrium analysis was performed at 4°C and a rotor speed of 9,000 rpm.
Figure 6
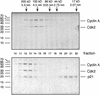
p21/Cyclin A/Cdk2 complexes are kinase inactive. (A) Purified cyclin A/Cdk2 complexes (lane 1) and p21/cyclin A/Cdk2 complexes (lane 2) were separated by SDS-PAGE and stained with Coomassie brilliant blue to estimate relative concentrations of their polypeptides (A). Size markers are shown at the sides. The kinase activities of diluted samples (1:100) were determined using [γ-32P] ATP and histone H1 as substrates. The amount of 32P incorporation in the histone H1 bands is shown after SDS-PAGE by autoradiography (bottom) (B) and determined using a PhosphorImager (C). Kinase activities are shown as percentage of that of the noninhibited complexes and were normalized for the kinase subunits present in the complexes. (C).
Figure 7
Inhibition by p21 is lost when inhibited kinase complexes were diluted under a threshold level. The complexes shown in Fig. 5A were diluted 1:1000 (left) and 1:10,000 (right). Kinase activities of p21-bound cyclin A/Cdk2 (+) were compared to that of noninhibited kinase (−) using [γ-32P] ATP and histone H1 as substrates. (A) The amount of 32P incorporation in the histone H1 bands is shown after SDS-PAGE by autoradiography (A). Two different exposures of the same gel are shown. The exposure time shown for the 10-fold diluted samples (1:10,000) is 10 times longer than that for the concentrated sample (1:1000). (B) Histone H1 kinase activities of all samples were determined using a PhosphorImager and the kinase activities of p21-inhibited kinase are shown as a percentage of that of noninhibited kinase and are normalized to the relative kinase subunit concentrations (B).
Figure 8
Phosphorylated p21 is a potent Cdk inhibitor. Phosphorylated p21 was obtained by incubating the inhibitor with an excess of activated cyclin A/Cdk2 and 1 m
m
ATP. Phosphorylated (p21-P) and mock-incubated, nonphosphorylated p21 (p21) were purified and immobilized on protein A–Sepharose beads as described in Fig. 2. These beads and anti-cyclin A antibodies bound to control beads were incubated with 250 ng of active cyclin A/Cdk2 complex and washed and equal aliquots were analyzed for associated kinase activities (histone H1 kinase) and the amount of precipitated proteins. Precipitated proteins were detected after SDS-PAGE and Western blotting and analyzed for precipitated kinase subunit Cdk2 (Cdk2, α-PSTAIRE) and inhibitor p21 (p21, α-p21). The phosphorylated forms of p21 show a lower electrophoretic mobility compared to nonmodified p21. Precipitated kinase activities associated with these samples were determined in assays using [γ-32P]ATP and histone H1 as substrate and detected after SDS-PAGE by autoradiography of the dried gel (histone H1 kinase).
Similar articles
- Cytoplasmic displacement of cyclin E-cdk2 inhibitors p21Cip1 and p27Kip1 in anchorage-independent cells.
Orend G, Hunter T, Ruoslahti E. Orend G, et al. Oncogene. 1998 May;16(20):2575-83. doi: 10.1038/sj.onc.1201791. Oncogene. 1998. PMID: 9632134 - Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver.
Albrecht JH, Poon RY, Ahonen CL, Rieland BM, Deng C, Crary GS. Albrecht JH, et al. Oncogene. 1998 Apr 23;16(16):2141-50. doi: 10.1038/sj.onc.1201728. Oncogene. 1998. PMID: 9572495 - The cell cycle inhibitor p21CIP is phosphorylated by cyclin A-CDK2 complexes.
Jaumot M, Estañol JM, Casanovas O, Graña X, Agell N, Bachs O. Jaumot M, et al. Biochem Biophys Res Commun. 1997 Dec 18;241(2):434-8. doi: 10.1006/bbrc.1997.7787. Biochem Biophys Res Commun. 1997. PMID: 9425288 - p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity.
Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Dorée M, Messier H, Fotedar A. Fotedar R, et al. Oncogene. 1996 May 16;12(10):2155-64. Oncogene. 1996. PMID: 8668341 - A new pathway for mitogen-dependent cdk2 regulation uncovered in p27(Kip1)-deficient cells.
Coats S, Whyte P, Fero ML, Lacy S, Chung G, Randel E, Firpo E, Roberts JM. Coats S, et al. Curr Biol. 1999 Feb 25;9(4):163-73. doi: 10.1016/s0960-9822(99)80086-4. Curr Biol. 1999. PMID: 10074425
Cited by
- Retinoic acid signaling and neuronal differentiation.
Janesick A, Wu SC, Blumberg B. Janesick A, et al. Cell Mol Life Sci. 2015 Apr;72(8):1559-76. doi: 10.1007/s00018-014-1815-9. Epub 2015 Jan 6. Cell Mol Life Sci. 2015. PMID: 25558812 Free PMC article. Review. - The p27-Skp2 axis mediates glucocorticoid-induced cell cycle arrest in T-lymphoma cells.
Kullmann MK, Grubbauer C, Goetsch K, Jäkel H, Podmirseg SR, Trockenbacher A, Ploner C, Cato AC, Weiss C, Kofler R, Hengst L. Kullmann MK, et al. Cell Cycle. 2013 Aug 15;12(16):2625-35. doi: 10.4161/cc.25622. Epub 2013 Jul 9. Cell Cycle. 2013. PMID: 23907123 Free PMC article. - Role of Cdk1 in the p53-independent abrogation of the postmitotic checkpoint by human papillomavirus E6.
Zhang W, Liu Y, Zhao N, Chen H, Qiao L, Zhao W, Chen JJ. Zhang W, et al. J Virol. 2015 Mar;89(5):2553-62. doi: 10.1128/JVI.02269-14. Epub 2014 Dec 17. J Virol. 2015. PMID: 25520504 Free PMC article. - Polyphenols as key players for the antileukaemic effects of propolis.
Abubakar MB, Abdullah WZ, Sulaiman SA, Ang BS. Abubakar MB, et al. Evid Based Complement Alternat Med. 2014;2014:371730. doi: 10.1155/2014/371730. Epub 2014 Mar 19. Evid Based Complement Alternat Med. 2014. PMID: 24772179 Free PMC article. Review. - A review of molecular mechanisms of the anti-leukemic effects of phenolic compounds in honey.
Abubakar MB, Abdullah WZ, Sulaiman SA, Suen AB. Abubakar MB, et al. Int J Mol Sci. 2012 Nov 15;13(11):15054-73. doi: 10.3390/ijms131115054. Int J Mol Sci. 2012. PMID: 23203111 Free PMC article. Review.
References
- Andrews P. Estimation of molecular size and molecular weights of biological compounds by gel filtration. Meth Biochem Anal. 1970;18:1–53. - PubMed
- Carnero A, Hannon G. The INK4 family of CDK inhibitors. Curr Top Microbiol Immunol. 1998;227:43–55. - PubMed
- Chen J, Jackson PK, Kirschner MW, Dutta A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature. 1995;374:386–388. - PubMed
- Harper J. Cyclin dependent kinase inhibitors. Cancer Surv. 1997;29:91–107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources