Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva - PubMed (original) (raw)
Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva
D K Mercer et al. Appl Environ Microbiol. 1999 Jan.
Abstract
Competitive PCR was used to monitor the survival of a 520-bp DNA target sequence from a recombinant plasmid, pVACMC1, after admixture of the plasmid with freshly sampled human saliva. The fraction of the target remaining amplifiable ranged from 40 to 65% after 10 min of exposure to saliva samples from five subjects and from 6 to 25% after 60 min of exposure. pVACMC1 plasmid DNA that had been exposed to degradation by fresh saliva was capable of transforming naturally competent Streptococcus gordonii DL1 to erythromycin resistance, although transforming activity decreased rapidly, with a half-life of approximately 50 s. S. gordonii DL1 transformants were obtained in the presence of filter-sterilized saliva and a 1-microg/ml final concentration of pVACMC1 DNA. Addition of filter-sterilized saliva instead of heat-inactivated horse serum to S. gordonii DL1 cells induced competence, although with slightly lower efficiency. These findings indicate that DNA released from bacteria or food sources within the mouth has the potential to transform naturally competent oral bacteria. However, further investigations are needed to establish whether transformation of oral bacteria can occur at significant frequencies in vivo.
Figures
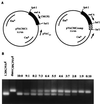
FIG. 1
Construction of the competitor plasmid to pVACMC1 and its use in competitive PCR. (A) Plasmid pVACMC1 and its competitor, pVACMCcomp, obtained by insertion of a 100-bp _Sal_I fragment (see text). (B) Trial competitive PCR experiment in which different ratios of competitor plasmid and pVACMC1 were subjected to PCR amplification with the forward PCR primer CMCP2 and the reverse primer pVACrev. The R. flavefaciens cellulase gene, endA, encodes CMCase activity.
FIG. 2
Persistence of a 520-bp fragment of pVACMC1 DNA after incubation with fresh, whole saliva for 9 min. PCR amplification of degraded DNA was carried out as described in Materials and Methods, and products were analyzed on a 1.4% (wt/vol) agarose gel. A 1-kb DNA ladder (Gibco BRL), 0.5 to 12.2 kbp, was used.
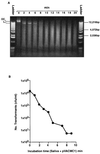
FIG. 3
(A) Survival of pVACMC1 DNA in human saliva analyzed on a 0.8% (wt/vol) agarose gel. A 1-kb DNA ladder (Gibco BRL), 0.5 to 12.2 kbp, was used. OC, open circular; L, linear. A faint band of covalently closed circular DNA was visible in lane 0). (B) Transformation of S. gordonii DL1 with pVACMC1 DNA that had been previously exposed to fresh, whole saliva for the times indicated.
FIG. 4
Transformation of S. gordonii DL1 with pVACMC1 in the presence and absence of filter-sterilized saliva. Competent cells were exposed to DNA and saliva or sterile dH2O for the times shown before plating on selective medium to identify transformants. Maximum transformation frequencies for the saliva of a second volunteer were 3.7 × 105. Experiments were carried out with different samples of S. gordonii DL1 competent cells.
FIG. 5
Transformation of S. gordonii DL1 with pVACMC1 after competence induction by different growth supplements. The transformation protocol described in Materials and Methods was followed except that heat-inactivated horse serum was replaced with the other growth supplements.
Similar articles
- Marker rescue studies of the transfer of recombinant DNA to Streptococcus gordonii in vitro, in foods and gnotobiotic rats.
Kharazmi M, Sczesny S, Blaut M, Hammes WP, Hertel C. Kharazmi M, et al. Appl Environ Microbiol. 2003 Oct;69(10):6121-7. doi: 10.1128/AEM.69.10.6121-6127.2003. Appl Environ Microbiol. 2003. PMID: 14532070 Free PMC article. - Transformation of an oral bacterium via chromosomal integration of free DNA in the presence of human saliva.
Mercer DK, Scott KP, Melville CM, Glover LA, Flint HJ. Mercer DK, et al. FEMS Microbiol Lett. 2001 Jun 25;200(2):163-7. doi: 10.1111/j.1574-6968.2001.tb10709.x. FEMS Microbiol Lett. 2001. PMID: 11425469 - Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro.
Aspiras MB, Kazmerzak KM, Kolenbrander PE, McNab R, Hardegen N, Jenkinson HF. Aspiras MB, et al. Appl Environ Microbiol. 2000 Sep;66(9):4074-83. doi: 10.1128/AEM.66.9.4074-4083.2000. Appl Environ Microbiol. 2000. PMID: 10966431 Free PMC article. - Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces.
Jenkinson HF, Terry SD, McNab R, Tannock GW. Jenkinson HF, et al. Infect Immun. 1993 Aug;61(8):3199-208. doi: 10.1128/iai.61.8.3199-3208.1993. Infect Immun. 1993. PMID: 8335350 Free PMC article. - Genetic competence and transformation in oral streptococci.
Cvitkovitch DG. Cvitkovitch DG. Crit Rev Oral Biol Med. 2001;12(3):217-43. doi: 10.1177/10454411010120030201. Crit Rev Oral Biol Med. 2001. PMID: 11497374 Review.
Cited by
- Marker rescue studies of the transfer of recombinant DNA to Streptococcus gordonii in vitro, in foods and gnotobiotic rats.
Kharazmi M, Sczesny S, Blaut M, Hammes WP, Hertel C. Kharazmi M, et al. Appl Environ Microbiol. 2003 Oct;69(10):6121-7. doi: 10.1128/AEM.69.10.6121-6127.2003. Appl Environ Microbiol. 2003. PMID: 14532070 Free PMC article. - Bacterial sex in dental plaque.
Olsen I, Tribble GD, Fiehn NE, Wang BY. Olsen I, et al. J Oral Microbiol. 2013 Jun 3;5. doi: 10.3402/jom.v5i0.20736. Print 2013. J Oral Microbiol. 2013. PMID: 23741559 Free PMC article. - Stress-Induced, Highly Efficient, Donor Cell-Dependent Cell-to-Cell Natural Transformation in Bacillus subtilis.
Zhang X, Jin T, Deng L, Wang C, Zhang Y, Chen X. Zhang X, et al. J Bacteriol. 2018 Aug 10;200(17):e00267-18. doi: 10.1128/JB.00267-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29941421 Free PMC article. - Various pathways leading to the acquisition of antibiotic resistance by natural transformation.
Domingues S, Nielsen KM, da Silva GJ. Domingues S, et al. Mob Genet Elements. 2012 Nov 1;2(6):257-260. doi: 10.4161/mge.23089. Mob Genet Elements. 2012. PMID: 23482877 Free PMC article. - Microbial analysis of bite marks by sequence comparison of streptococcal DNA.
Kennedy DM, Stanton JA, García JA, Mason C, Rand CJ, Kieser JA, Tompkins GR. Kennedy DM, et al. PLoS One. 2012;7(12):e51757. doi: 10.1371/journal.pone.0051757. Epub 2012 Dec 19. PLoS One. 2012. PMID: 23284761 Free PMC article.
References
- Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182:490–497. - PubMed
- Duval-Iflah Y, Gainche I, Ouriet M-F, Lett M-C, Hubert J-C. Recombinant DNA transfer to Escherichia coli of human faecal origin in vitro and in digestive tract of gnotobiotic mice. FEMS Microbiol Ecol. 1994;15:79–88.
- Ehrlich S D. Biotechnology action programme (Biotechnology R & D in the EC, part II). Commission of the European Communities catalogue of BAP achievements on risk assessment for the period 1985–1990. Brussels, Belgium: Cordis-RTD Publications; 1990. Gene transfer from and survival of genetically modified lactic acid bacteria; pp. 1–91.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources