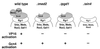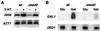Mediator protein mutations that selectively abolish activated transcription - PubMed (original) (raw)
Mediator protein mutations that selectively abolish activated transcription
L C Myers et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Deletion of any one of three subunits of the yeast Mediator of transcriptional regulation, Med2, Pgd1 (Hrs1), and Sin4, abolished activation by Gal4-VP16 in vitro. By contrast, other Mediator functions, stimulation of basal transcription and of TFIIH kinase activity, were unaffected. A different but overlapping Mediator subunit dependence was found for activation by Gcn4. The genetic requirements for activation in vivo were closely coincident with those in vitro. A whole genome expression profile of a Deltamed2 strain showed diminished transcription of a subset of inducible genes but only minor effects on "basal" transcription. These findings make an important connection between transcriptional activation in vitro and in vivo, and identify Mediator as a "global" transcriptional coactivator.
Figures
Figure 1
Transcription assays of wild-type and Δpgd1 holoenzymes. Transcription was performed with highly purified transcription factors and DNA templates containing binding sites for Gcn4 (GCN4:G−) and Gal4 (GAL4:G−). Gal4–VP16 activation (31-fold for wild-type holoenzyme, 1.8-fold for Δpgd1) was quantitated by comparing transcription in the presence and absence of the activator on the GAL4:G− template and dividing the ratio by any change in transcription of GCN4:G− template. Gcn4 activation (8.2-fold for wild-type holoenzyme, 6.9-fold for Δpgd1) was measured with the GCN4:G− template in a similar manner.
Figure 2
Immunobloting analysis of wild-type and mutant holoenzymes. Mono-Q fractions of wild-type, Δmed2, and Δpgd1 holoenzymes were subjected to immunoblot analysis by using antibodies directed against Mediator components Med2, Pgd1, Med4, and Med7 (8). The amounts of the Δmed2 and Δpgd1 holoenzymes loaded on the gel were approximately three times greater than the amount of wild-type holoenzyme, to demonstrate the absence of Med2 and Pgd1 subunits.
Figure 3
Structure–function relationships of wild-type and mutant RNA polymerase II holoenzymes. The subunit organization of the Sin4/Rgr1 module of Mediator is based on Fig. 2 and the Results in the text. This model, however, does not preclude the existence of weak interactions among Med2, Pgd1, Sin4, and other subunits of holopolymerase that do not withstand the rigors of purification. The functional consequences of the various Mediator mutations are from Table 1.
Figure 4
RNA blot analysis of HIS4 and GAL1 induction in wild-type and Δmed2 strains. (A) Wild-type (MG106) and Δmed2 (MG107) mutant cells transformed with pRS313 (HIS3) (22) were grown in synthetic minimal medium (16) supplemented with 0.2 mM inositol, 2.0 mM leucine, 0.5 mM isoleucine, 0.5 mM valine, 0.4 mM tryptophan, 0.25 mM arginine, 0.1 mM adenine, 0.2 mM lysine, and 0.2 mM uracil to OD600 = 0.8. For starvation conditions, 3-aminotriazol (3-AT) was added to 100 mM, and the cultures were harvested 3 hr later. The RNA blot was hybridized to radioactively labeled probes for HIS4 and ACT1. (B) Wild-type (MG106) and Δmed2 (MG107) mutant cells were grown in yeast extract/peptone/raffinose medium overnight, washed with water and transferred to yeast extract/peptone/glucose (Glu) or yeast extract/peptone/galactose (Gal) media at a density of OD600 = 0.15, followed by harvest at OD600 = 0.6. The RNA blot was hybridized to radioactively labeled probes for GAL1 and DED1.
Figure 5
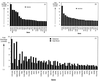
Genes that display defects in transcription under galactose and heat shock growth conditions in a Δmed2 strain. Genes are identified by gene name or ORF designation (as listed in the Stanford Genome Database). (A) The 22 genes that suffer the greatest transcription defects in the Δmed2 strain under galactose induction conditions are shown. The GAL genes shown in Fig. 6_A_ are not included in this plot. (B) The 22 genes that suffer the greatest transcription defects in the Δmed2 strain under heat shock growth conditions (excluding genes described in Fig. 6_B_) are shown. (C) All genes that suffer a >2-fold defect in the Δmed2 strain under both galactose and heat shock growth conditions are shown. The deleted gene, MED2, was not included on the above lists. “Fold Decrease (Δmed2)” refers to the ratio (wild type/Δmed2) of normalized transcript levels.
Figure 6
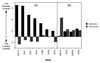
Galactose- and heat shock-induced genes that display transcription defects in a Δmed2 strain. (A) Well characterized galactose-induced genes that suffer a >2-fold defect in expression in the Δmed2 strain grown on galactose medium are shown. For comparison, fold differences in expression of these same genes under heat shock conditions (in glucose) also are indicated. (B) Well characterized heat shock-induced genes that suffer a >2-fold defect in expression in the Δmed2 strain under heat shock (37°C) growth conditions are shown. For comparison, fold differences in expression of these same genes under galactose growth conditions (30°C) also are indicated. “Fold Decrease (Δmed2)” and “Fold Increase (Δmed2)” refer to ratios (wild-type/Δmed2 and Δmed2/wild-type, respectively) of normalized transcript levels.
Comment in
- Transcriptional regulation: contending with complexity.
Holstege FC, Young RA. Holstege FC, et al. Proc Natl Acad Sci U S A. 1999 Jan 5;96(1):2-4. doi: 10.1073/pnas.96.1.2. Proc Natl Acad Sci U S A. 1999. PMID: 9874759 Free PMC article. No abstract available.
Similar articles
- Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation.
Reeves WM, Hahn S. Reeves WM, et al. Mol Cell Biol. 2003 Jan;23(1):349-58. doi: 10.1128/MCB.23.1.349-358.2003. Mol Cell Biol. 2003. PMID: 12482986 Free PMC article. - A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II.
Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. Kim YJ, et al. Cell. 1994 May 20;77(4):599-608. doi: 10.1016/0092-8674(94)90221-6. Cell. 1994. PMID: 8187178 - Genetic characterization of rbt mutants that enhance basal transcription from core promoters in Saccharomyces cerevisiae.
Kunoh T, Sakuno T, Furukawa T, Kaneko Y, Harashima S. Kunoh T, et al. J Biochem. 2000 Oct;128(4):575-84. doi: 10.1093/oxfordjournals.jbchem.a022789. J Biochem. 2000. PMID: 11011139 - Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo.
Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Kuo MH, et al. Genes Dev. 1998 Mar 1;12(5):627-39. doi: 10.1101/gad.12.5.627. Genes Dev. 1998. PMID: 9499399 Free PMC article. - Mediator of transcriptional regulation.
Björklund S, Kim YJ. Björklund S, et al. Trends Biochem Sci. 1996 Sep;21(9):335-7. doi: 10.1016/s0968-0004(96)10051-7. Trends Biochem Sci. 1996. PMID: 8870496 Review.
Cited by
- The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II.
Bhoite LT, Yu Y, Stillman DJ. Bhoite LT, et al. Genes Dev. 2001 Sep 15;15(18):2457-69. doi: 10.1101/gad.921601. Genes Dev. 2001. PMID: 11562354 Free PMC article. - Structural organization of yeast and mammalian mediator complexes.
Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Dotson MR, et al. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14307-10. doi: 10.1073/pnas.260489497. Proc Natl Acad Sci U S A. 2000. PMID: 11114191 Free PMC article. - Mediator subunits and histone methyltransferase Set2 contribute to Ino2-dependent transcriptional activation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae.
Dettmann A, Jäschke Y, Triebel I, Bogs J, Schröder I, Schüller HJ. Dettmann A, et al. Mol Genet Genomics. 2010 Mar;283(3):211-21. doi: 10.1007/s00438-009-0508-9. Mol Genet Genomics. 2010. PMID: 20054697 - Repression and activation domains of RME1p structurally overlap, but differ in genetic requirements.
Blumental-Perry A, Li W, Simchen G, Mitchell AP. Blumental-Perry A, et al. Mol Biol Cell. 2002 May;13(5):1709-21. doi: 10.1091/mbc.01-09-0468. Mol Biol Cell. 2002. PMID: 12006664 Free PMC article. - Malleable machines in transcription regulation: the mediator complex.
Tóth-Petróczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. Tóth-Petróczy A, et al. PLoS Comput Biol. 2008 Dec;4(12):e1000243. doi: 10.1371/journal.pcbi.1000243. Epub 2008 Dec 19. PLoS Comput Biol. 2008. PMID: 19096501 Free PMC article.
References
- Kelleher R J, III, Flanagan P M, Kornberg R D. Cell. 1990;61:1209–1216. - PubMed
- Flanagan P M, Kelleher R J, III, Sayre M H, Tschochner H, Kornberg R D. Nature (London) 1991;350:436–438. - PubMed
- Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. - PubMed
- Koleske A J, Young R A. Nature (London) 1994;368:466–469. - PubMed
- Carlson M. Annu Rev Cell Dev Biol. 1997;13:1–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials