Septal localization of FtsQ, an essential cell division protein in Escherichia coli - PubMed (original) (raw)
Septal localization of FtsQ, an essential cell division protein in Escherichia coli
J C Chen et al. J Bacteriol. 1999 Jan.
Abstract
Septation in Escherichia coli requires several gene products. One of these, FtsQ, is a simple bitopic membrane protein with a short cytoplasmic N terminus, a membrane-spanning segment, and a periplasmic domain. We have constructed a merodiploid strain that expresses both FtsQ and the fusion protein green fluorescent protein (GFP)-FtsQ from single-copy chromosomal genes. The gfp-ftsQ gene complements a null mutation in ftsQ. Fluorescence microscopy revealed that GFP-FtsQ localizes to the division site. Replacing the cytoplasmic and transmembrane domains of FtsQ with alternative membrane anchors did not prevent the localization of the GFP fusion protein, while replacing the periplasmic domain did, suggesting that the periplasmic domain is necessary and sufficient for septal targeting. GFP-FtsQ localization to the septum depended on the cell division proteins FtsZ and FtsA, which are cytoplasmic, but not on FtsL and FtsI, which are bitopic membrane proteins with comparatively large periplasmic domains. In addition, the septal localization of ZipA apparently did not require functional FtsQ. Our results indicate that FtsQ is an intermediate recruit to the division site.
Figures
FIG. 1
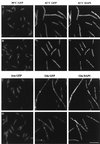
Subcellular locations of GFP-FtsQ (A), GFP-FFQ (B), and GFP (C). Cells were grown and prepared for microscopy as described in Materials and Methods. Strains used were EC442 (A), JOE196 (B), and EC452 (C). Bar, 10 μm.
FIG. 2
Immunoblot of FtsQ and GFP-FtsQ fusion proteins. Lanes: 1, MC4100; 2 to 6, serial dilutions of EC442; 7, MC4100; 8, JOE192; 9, JOE193; 10, JOE196; 11, JOE194. Cells were grown at 30°C to the early log phase and boiled in SDS sample buffer as described in Materials and Methods. Equivalent amounts of samples were loaded except that for lanes 3 to 6, the sample in lane 2 was diluted 1/5, 1/10, 1/15, and 1/20, respectively, with SDS sample buffer. The positions of GFP-FtsQ and FtsQ are indicated on the right.
FIG. 3
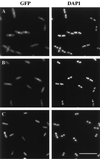
Localization of GFP-FtsQ in fts mutants. (A and B) GFP-FtsQ localizes to division sites in ftsZ84(Ts) (A) and ftsA12(Ts) (B) cells at 30°C (left panels) but not at 42°C (middle panels); DAPI images indicate that nucleoid structures are normal in the filaments at 42°C (right panels). (C and D) GFP-FtsQ localizes to potential division sites in FtsI (C) and FtsL (D) depletion strains; the left panels are GFP images of cells growing normally in the presence of arabinose, the middle panels are GFP images of cells grown with glucose and therefore depleted of FtsI or FtsL, and the right panels are corresponding DAPI images of the middle panels. Cells were grown and fixed for fluorescence microscopy as described in Materials and Methods. Strains used were JOE97 (A), JOE95 (B), JOE233 (C), and JOE220 (D). Bar, 10 μm.
FIG. 4
Localization of ZipA-GFP in an ftsQ1(Ts) mutant. (Left) GFP image of JOE165 cells grown at 30°C. (Right) GFP image of JOE165 cells grown at 42°C for 45 min. Cells were fixed for microscopy as described in Materials and Methods. Bar, 10 μm.
Similar articles
- Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL.
Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Weiss DS, et al. J Bacteriol. 1999 Jan;181(2):508-20. doi: 10.1128/JB.181.2.508-520.1999. J Bacteriol. 1999. PMID: 9882665 Free PMC article. - Localization of FtsL to the Escherichia coli septal ring.
Ghigo JM, Weiss DS, Chen JC, Yarrow JC, Beckwith J. Ghigo JM, et al. Mol Microbiol. 1999 Jan;31(2):725-37. doi: 10.1046/j.1365-2958.1999.01213.x. Mol Microbiol. 1999. PMID: 10027987 - FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division.
Chen JC, Beckwith J. Chen JC, et al. Mol Microbiol. 2001 Oct;42(2):395-413. doi: 10.1046/j.1365-2958.2001.02640.x. Mol Microbiol. 2001. PMID: 11703663 - Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli.
Guzman LM, Weiss DS, Beckwith J. Guzman LM, et al. J Bacteriol. 1997 Aug;179(16):5094-103. doi: 10.1128/jb.179.16.5094-5103.1997. J Bacteriol. 1997. PMID: 9260951 Free PMC article. - Contribution of the FtsQ transmembrane segment to localization to the cell division site.
Scheffers DJ, Robichon C, Haan GJ, den Blaauwen T, Koningstein G, van Bloois E, Beckwith J, Luirink J. Scheffers DJ, et al. J Bacteriol. 2007 Oct;189(20):7273-80. doi: 10.1128/JB.00723-07. Epub 2007 Aug 10. J Bacteriol. 2007. PMID: 17693520 Free PMC article.
Cited by
- Coevolution with bacteriophages drives genome-wide host evolution and constrains the acquisition of abiotic-beneficial mutations.
Scanlan PD, Hall AR, Blackshields G, Friman VP, Davis MR Jr, Goldberg JB, Buckling A. Scanlan PD, et al. Mol Biol Evol. 2015 Jun;32(6):1425-35. doi: 10.1093/molbev/msv032. Epub 2015 Feb 12. Mol Biol Evol. 2015. PMID: 25681383 Free PMC article. - Crystal structure of the cell division protein FtsA from Thermotoga maritima.
van den Ent F, Löwe J. van den Ent F, et al. EMBO J. 2000 Oct 16;19(20):5300-7. doi: 10.1093/emboj/19.20.5300. EMBO J. 2000. PMID: 11032797 Free PMC article. - Evidence for polar positional information independent of cell division and nucleoid occlusion.
Janakiraman A, Goldberg MB. Janakiraman A, et al. Proc Natl Acad Sci U S A. 2004 Jan 20;101(3):835-40. doi: 10.1073/pnas.0305747101. Epub 2004 Jan 8. Proc Natl Acad Sci U S A. 2004. PMID: 14715903 Free PMC article. - Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK.
Geissler B, Margolin W. Geissler B, et al. Mol Microbiol. 2005 Oct;58(2):596-612. doi: 10.1111/j.1365-2958.2005.04858.x. Mol Microbiol. 2005. PMID: 16194242 Free PMC article. - Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery.
Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Chung HS, et al. Proc Natl Acad Sci U S A. 2009 Dec 22;106(51):21872-7. doi: 10.1073/pnas.0911674106. Epub 2009 Dec 7. Proc Natl Acad Sci U S A. 2009. PMID: 19995973 Free PMC article.
References
- Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. - PubMed
- Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
- Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. Unpublished data.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases