Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli - PubMed (original) (raw)
Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli
K De Pina et al. J Bacteriol. 1999 Jan.
Abstract
Expression of the nickel-specific transport system encoded by the Escherichia coli nikABCDE operon is repressed by a high concentration of nickel. By using random transposon Tn10 insertion, we isolated mutants in which expression of the nik operon became constitutive with respect to nickel. We have identified the corresponding nikR gene which encodes a nickel-responsive regulator. Expression of nikR was partially controlled by Fnr through transcription from the nikA promoter region. In addition, a specific transcription start site for the constitutive expression of nikR was found 51 bp upstream of the nikR gene.
Figures
FIG. 1
Complementation analysis of Tn_10_ insertion mutants with plasmids carrying the nik region. KS01 and KS02 are Tn_10_ insertion derivatives of HYD723 (nikA-lacZ). Cells which carried the indicated plasmid were grown microaerobically at 37°C in LB medium supplemented with 2 μM ammonium molybdate, 2 μM sodium selenite, and kanamycin (25 μg/ml) when required (21). Plasmids pLW22, pLW25, and pLW26 were described previously (24). β-Galactosidase activity was measured for cells treated by addition of 0.0025% sodium dodecyl sulfate–5% chloroform, and the specific activity is expressed as nanomoles of _o_-nitrophenol produced per minute per milligram bacterial dry weight. Values quoted are the averages of three separate experiments. Symbols: E, _Eco_RI; H, _Hin_cII; M, _Mlu_I; N, _Nsi_I; S, _Ssp_I; V, _Eco_RV.
FIG. 2
Specific expression of the nikR, nikD, and nikE gene products, under control of the T7 φ10 promoter in E. coli K38/pGP1-2. Cells containing vector pT7-6 (lane 1), its derivative pHD4, harboring nikDER (lane 2), vector pKSM710 (lane 4), and its derivative p8611, harboring nikR (lane 3), were labeled with [35S]methionine and [35S]cysteine and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 17% denaturing polyacrylamide gel. Molecular mass standards (in kilodaltons) and NikE, NikD, and NikR proteins are indicated on the right and on the left, respectively.
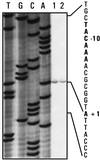
FIG. 3
Determination of the constitutive nikR transcription start site. Total RNAs (50 μg) isolated from E. coli NM522/pLW22 grown anaerobically in Luria broth in the absence (lane 1) or presence (lane 2) of 0.5 mM NiCl2 were analyzed by primer extension with primer NikR (see text). The DNA sequence ladder (lanes TGCA) was obtained with the same primer and plasmid pLW22 as a template. Part of the sequence and the nikR transcription start site are indicated on the right.
Similar articles
- Specific metal recognition in nickel trafficking.
Higgins KA, Carr CE, Maroney MJ. Higgins KA, et al. Biochemistry. 2012 Oct 9;51(40):7816-32. doi: 10.1021/bi300981m. Epub 2012 Sep 28. Biochemistry. 2012. PMID: 22970729 Free PMC article. Review. - Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity.
Jubier-Maurin V, Rodrigue A, Ouahrani-Bettache S, Layssac M, Mandrand-Berthelot MA, Köhler S, Liautard JP. Jubier-Maurin V, et al. J Bacteriol. 2001 Jan;183(2):426-34. doi: 10.1128/JB.183.2.426-434.2001. J Bacteriol. 2001. PMID: 11133934 Free PMC article. - Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori.
Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Contreras M, et al. Mol Microbiol. 2003 Aug;49(4):947-63. doi: 10.1046/j.1365-2958.2003.03621.x. Mol Microbiol. 2003. PMID: 12890020 - Regulation of high affinity nickel uptake in bacteria. Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites.
Chivers PT, Sauer RT. Chivers PT, et al. J Biol Chem. 2000 Jun 30;275(26):19735-41. doi: 10.1074/jbc.M002232200. J Biol Chem. 2000. PMID: 10787413 - FNR and its role in oxygen-regulated gene expression in Escherichia coli.
Spiro S, Guest JR. Spiro S, et al. FEMS Microbiol Rev. 1990 Aug;6(4):399-428. doi: 10.1111/j.1574-6968.1990.tb04109.x. FEMS Microbiol Rev. 1990. PMID: 2248796 Review.
Cited by
- Specific metal recognition in nickel trafficking.
Higgins KA, Carr CE, Maroney MJ. Higgins KA, et al. Biochemistry. 2012 Oct 9;51(40):7816-32. doi: 10.1021/bi300981m. Epub 2012 Sep 28. Biochemistry. 2012. PMID: 22970729 Free PMC article. Review. - Environmental factors affecting the expression of pilAB as well as the proteome and transcriptome of the grass endophyte Azoarcus sp. strain BH72.
Hauberg-Lotte L, Klingenberg H, Scharf C, Böhm M, Plessl J, Friedrich F, Völker U, Becker A, Reinhold-Hurek B. Hauberg-Lotte L, et al. PLoS One. 2012;7(1):e30421. doi: 10.1371/journal.pone.0030421. Epub 2012 Jan 20. PLoS One. 2012. PMID: 22276194 Free PMC article. - NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori.
van Vliet AH, Poppelaars SW, Davies BJ, Stoof J, Bereswill S, Kist M, Penn CW, Kuipers EJ, Kusters JG. van Vliet AH, et al. Infect Immun. 2002 Jun;70(6):2846-52. doi: 10.1128/IAI.70.6.2846-2852.2002. Infect Immun. 2002. PMID: 12010971 Free PMC article. - Acid-responsive gene induction of ammonia-producing enzymes in Helicobacter pylori is mediated via a metal-responsive repressor cascade.
van Vliet AH, Kuipers EJ, Stoof J, Poppelaars SW, Kusters JG. van Vliet AH, et al. Infect Immun. 2004 Feb;72(2):766-73. doi: 10.1128/IAI.72.2.766-773.2004. Infect Immun. 2004. PMID: 14742519 Free PMC article. - Form and function in metal-dependent transcriptional regulation: dawn of the enlightenment.
Rensing C. Rensing C. J Bacteriol. 2005 Jun;187(12):3909-12. doi: 10.1128/JB.187.12.3909-3912.2005. J Bacteriol. 2005. PMID: 15937153 Free PMC article. No abstract available.
References
- Babich H, Stotzky G. Toxicity of nickel to microbes: environmental aspects. Adv Appl Microbiol. 1983;29:195–265. - PubMed
- Bardonnet N, Blanco C. ′uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol Lett. 1992;72:243–247. - PubMed
- de Pina K, Navarro C, McWalter L, Boxer D H, Price N C, Kelly S M, Mandrand-Berthelot M A, Wu L F. Purification and characterization of the periplasmic nickel-binding protein NikA of Escherichia coli K12. Eur J Biochem. 1995;227:857–865. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous