Cytosine methylation and mammalian development - PubMed (original) (raw)
Cytosine methylation and mammalian development
C P Walsh et al. Genes Dev. 1999.
Abstract
Programmed methylation and demethylation of regulatory sequences has been proposed to play a central role in vertebrate development. We report here that the methylation status of the 5' regions of a panel of tissue-specific genes could not be correlated with expression in tissues of fetal and newborn mice. Genes reported to be regulated by reversible methylation were not expressed ectopically or precociously in Dnmt1-deficient mouse embryos under conditions where demethylation caused biallelic expression of imprinted genes and activated transcription of endogenous retroviruses of the IAP class. These and other data suggest that the numerous published expression-methylation correlations may have described not a cause but a consequence of transcriptional activation. A model is proposed under which cytosine methylation represents a biochemical specialization of large genomes that participates in specialized biological functions such as allele-specific gene expression and the heritable transcriptional silencing of parasitic sequence elements, whereas cellular differentiation is controlled by conserved regulatory networks that do not depend on covalent modification of the genome.
Figures
Figure 1
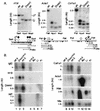
Methylation analysis of the 5′ regions of tissue-specific genes. Diamonds above the heavy line indicate _Hpa_II sites for all but E, in which _Hha_I sites (5′-GCGC-3′) are denoted. The amount of fill in the diamond symbol represents average methylation level in nonexpressing tissues. Tick marks below the heavy line indicate the positions of each CpG dinucleotide. Probes and predicted fragment sizes are shown at the bottom of each panel, and transcription start sites are indicated by arrows. Lanes C contained DNA not cleaved with methylation-sensitive enzymes; lanes M contained DNA digested with _Msp_I (absent in E, which contained _Hha_I digests). DNA of sperm is completely methylated at tested sites in A, B, G, and H and completely unmethylated in C, D, E, and F. Except for Lep in D, none of the latter genes show detectable methylation in somatic tissues. Prf1 in I is completely unmethylated in _Prf1_-expressing cytotoxic T lymphocytes and largely unmethylated in lung, liver, and whole E14.5 embryos, none of which express appreciable levels of Prf1. Methylation levels of non-CpG island genes are generally higher in brain and whole E11.5 embryos than in E14.5 embryos and adult organs. (E11.5 and E14.5) Whole mouse embryos at 11.5 and 14.5 days postcoitum; (Ki) kidney; (Br) brain; (Liv) liver; (Spl) spleen; (Mu) skeletal muscle from body wall; (Sp) sperm; (Ht) heart; (CTL) cytotoxic T cells purified by flow sorting mouse lymphocytes stained with antibody to CD8.
Figure 2
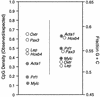
CpG densities predict methylation status of 5′ regions of tissue-specific genes in sperm DNA. The 5′ regions of the genes in Fig. 1 were analyzed for CpG density, G+C content, and methylation status. (•) Methylated sequences; (○) unmethylated sequences. Notice that sequences with higher observed/expected values for CpG densities tend to be unmethylated in sperm DNA. CpG densities (left) are a more accurate predictor of methylation status in sperm DNA than are G+C contents (right).
Figure 3
Normal expression of tissue-specific genes in DNA methyltransferase-deficient mouse embryos. (A) The genes of interest were largely unmethylated in control samples and underwent additional demethylation in Dnmt1-deficient mouse embryos at day E9.5. (B) A single RNA blot was hybridized consecutively with the eight probes shown. H19 and IAP retroviral transcripts can be seen to increase in the mutants; Igf2 declines and Dnmt1 mRNA is barely detectable. Col1a1, Acta1, and Hbb mRNAs are largely unaffected or reduced in amount in the mutant embryos, although all three genes had been reported to be repressed by cytosine methylation.
Similar articles
- Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing.
Gaudet F, Rideout WM 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Gaudet F, et al. Mol Cell Biol. 2004 Feb;24(4):1640-8. doi: 10.1128/MCB.24.4.1640-1648.2004. Mol Cell Biol. 2004. PMID: 14749379 Free PMC article. - Genetic Studies on Mammalian DNA Methyltransferases.
Dan J, Chen T. Dan J, et al. Adv Exp Med Biol. 2016;945:123-150. doi: 10.1007/978-3-319-43624-1_6. Adv Exp Med Biol. 2016. PMID: 27826837 Review. - H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19.
Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang SP, Pfeifer K. Srivastava M, et al. Genes Dev. 2000 May 15;14(10):1186-95. Genes Dev. 2000. PMID: 10817754 Free PMC article. - CpG hypomethylation in a large domain encompassing the embryonic beta-like globin genes in primitive erythrocytes.
Hsu M, Mabaera R, Lowrey CH, Martin DI, Fiering S. Hsu M, et al. Mol Cell Biol. 2007 Jul;27(13):5047-54. doi: 10.1128/MCB.02234-06. Epub 2007 Apr 23. Mol Cell Biol. 2007. PMID: 17452448 Free PMC article. - Role of Mammalian DNA Methyltransferases in Development.
Chen Z, Zhang Y. Chen Z, et al. Annu Rev Biochem. 2020 Jun 20;89:135-158. doi: 10.1146/annurev-biochem-103019-102815. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815535 Review.
Cited by
- Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos.
Stancheva I, Meehan RR. Stancheva I, et al. Genes Dev. 2000 Feb 1;14(3):313-27. Genes Dev. 2000. PMID: 10673503 Free PMC article. - Conserved plant genes with similarity to mammalian de novo DNA methyltransferases.
Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, Jacobsen SE. Cao X, et al. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4979-84. doi: 10.1073/pnas.97.9.4979. Proc Natl Acad Sci U S A. 2000. PMID: 10781108 Free PMC article. - The genome of the stick insect Medauroidea extradentata is strongly methylated within genes and repetitive DNA.
Krauss V, Eisenhardt C, Unger T. Krauss V, et al. PLoS One. 2009 Sep 29;4(9):e7223. doi: 10.1371/journal.pone.0007223. PLoS One. 2009. PMID: 19787064 Free PMC article. - Transcriptional regulation of mouse mast cell protease-2 by interleukin-15.
Mirghomizadeh F, Bullwinkel J, Orinska Z, Janssen O, Petersen A, Singh PB, Bulfone-Paus S. Mirghomizadeh F, et al. J Biol Chem. 2009 Nov 20;284(47):32635-41. doi: 10.1074/jbc.M109.015446. Epub 2009 Oct 1. J Biol Chem. 2009. PMID: 19801677 Free PMC article. - Effects of cytosine methylation on transcription factor binding sites.
Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H, Lassmann T, Harbers M, Forrest AR, Bajic VB; FANTOM consortium. Medvedeva YA, et al. BMC Genomics. 2014 Mar 26;15:119. doi: 10.1186/1471-2164-15-119. BMC Genomics. 2014. PMID: 24669864 Free PMC article.
References
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;6:503–514. - PubMed
- Baron MH, Maniatis T. Rapid reprogramming of globin gene expression in transient heterokaryons. Cell. 1986;46:591–602. - PubMed
- Barton PJ, Cohen L, Robert B, Fiszman MY, Bonhomme F, Guenet JL, Leader DP, Buckingham ME. The myosin alkali light chains of mouse ventricular and slow skeletal muscle are indistinguishable and are encoded by the same gene. J Biol Chem. 1985;260:8578–8584. - PubMed
- Bestor TH, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzyme is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. - PubMed
- Bestor TH. DNA methylation: How a bacterial immune function has evolved into a regulator of gene expression and genome structure in higher eukaryotes. Phil Trans R Soc Lond Biol Sci. 1990;326:179–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials