PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins - PubMed (original) (raw)
PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins
B Ren et al. Genes Dev. 1999.
Abstract
The PRDI-BF1/Blimp-1 protein is a transcriptional repressor required for normal B-cell differentiation, and it has been implicated in the repression of beta-interferon (IFN-beta) and c-myc gene expression. Here, we show that PRDI-BF1 represses transcription of the IFN-beta promoter and of an artificial promoter through an active repression mechanism. We also identified a minimal repression domain in PRDI-BF1 that is sufficient for transcriptional repression when tethered to DNA as a Gal4 fusion protein. Remarkably, this repression domain interacts specifically with hGrg, TLE1, and TLE2 proteins, all of which are members of the Groucho family of transcriptional corepressors. In addition, the hGrg protein itself can function as a potent repressor when tethered to DNA through the Gal4 DNA-binding domain. We also find that the amino-terminal glutamine-rich domains of hGrg and TLE1 are sufficient to mediate dimerization of the two Groucho family proteins. Proteins containing only this domain can function as a dominant-negative inhibitor of PRDI-BF1 repression, and can significantly increase the IFN-beta promoter activity after virus induction. We conclude that PRDI-BF1/Blimp-1 represses transcription by recruiting a complex of Groucho family proteins to DNA, and suggest that such corepressor complexes are required for the postinduction repression of the IFN-beta promoter.
Figures
Figure 1
PRDI-BF1 is an active repressor. (A) PRDI-BF1 represses transcription from the natural IFN-β promoter, and this repression requires the amino terminus of the protein. The histogram shows the CAT activity produced in HeLa cells transfected with 5 μg of the reporter gene containing the −110 IFN-β promoter fused to the CAT gene, cotransfected with 1 μg of the indicated expression vector, 4 μg of the pcDNA3 vector, and 2 μg of pCMV–lacZ. The cells were either untreated (dark bars) or infected with Sendai virus (open bars) 24 hr after transfection, and harvested 16 hr later. (Control) pcDNA3 vector. CAT activities in this and the following experiments are normalized to the activity of the cotransfected pCMV–lacZ gene. (B) PRDI-BF1 represses transcription of the tk promoter when bound to sites located 1000 nucleotides from the start site of transcription. HeLa cells were transfected with 2 μg of pCMV–lacZ control plasmid, 6 μg of pXM, 3 μg of reporter, and 1 μg of effector pcDNA3 (−) or pcDNA3–PRDI-BF1 (+). The control reporter BLCAT2 contains a fragment (−109 to +55 bp) of the herpes simplex virus tk promoter driving the expression of the bacterial CAT gene. The 4×PRF reporter contains four copies of the PRF element inserted adjacent to the tk promoter. In the case of the 4×PRF + 1.1-kb reporter a 1.1-kb λDNA fragment was inserted between the tk promoter and the PRF elements. The data shown are representative of three independent assays. For each reporter, the CAT activity for PRDI-BF1 was normalized to that of the negative control pcDNA3. (C) Amino acids 331–398 of PRDI-BF1 functions as a transcriptional repression domain. The PRDI-BF1 sequences tested in each assay are illustrated at left. Horizontal lines represent PRDI-BF1 sequences of the full-length protein and various deletion mutants as illustrated. Shaded boxes denote the zinc finger DNA-binding domains of PRDI-BF1. PRDI-BF1 constructs (1 μg of each) were transfected into HeLa cells with the same control reporter and PRF_-containing reporter (4×_PRF) as in B. The data shown are representative of three independent assays, and the CAT activity for all PRDI-BF1 constructs was normalized by the CAT activity of the cells transfected with the control pcDNA3–Flag vector and the respective reporter BLCAT2 (dark bars) or 4×PRF–BLCAT2 (open bars).
Figure 2
The PRDI-BF1 repression domain functions when fused to the Gal4-DNA-binding domain. (A) The ability of various Gal4–PRDI-BF1 fusion proteins to repress transcription was tested with a reporter that bears five Gal4 DNA-binding sites upstream of the tk promoter (G5BLCAT2). The BLCAT2 (see Fig. 1B) reporter was used as a control. Horizontal lines represent various PRDI-BF1 amino- and carboxy-terminal truncations cloned into the pBXG expression vector, which contains a DNA sequence encoding the Gal4 DNA-binding domain, Gal4(1–147) (boxed). PRDI-BF1 constructs (1 μg) were transfected into HeLa cells with either the control reporter BLCAT2 (dark bars) or G5BLCAT2 (open bars). The data shown are representative of three independent assays, and the CAT activities for all Gal4–PRDI-BF1 constructs were normalized by the CAT activity of the cells transfected with the control vector pECE and the respective reporter G5BLCAT2 or BLCAT2. (B) The sequence of a portion of the repression domain in PRDI-BF1. The sequence of the homologous region in Blimp-1 is also shown. The amino acid residues shared by the two proteins are listed between the two sequences. Two stretches of proline-rich regions, PRI and PRII, are indicated by brackets. (C) The proline-rich region of PRDI-BF1 is compared to similar region in WT1. Residues shared by PRDI-BF1 and WT1 protein are listed between the two sequences.
Figure 2
The PRDI-BF1 repression domain functions when fused to the Gal4-DNA-binding domain. (A) The ability of various Gal4–PRDI-BF1 fusion proteins to repress transcription was tested with a reporter that bears five Gal4 DNA-binding sites upstream of the tk promoter (G5BLCAT2). The BLCAT2 (see Fig. 1B) reporter was used as a control. Horizontal lines represent various PRDI-BF1 amino- and carboxy-terminal truncations cloned into the pBXG expression vector, which contains a DNA sequence encoding the Gal4 DNA-binding domain, Gal4(1–147) (boxed). PRDI-BF1 constructs (1 μg) were transfected into HeLa cells with either the control reporter BLCAT2 (dark bars) or G5BLCAT2 (open bars). The data shown are representative of three independent assays, and the CAT activities for all Gal4–PRDI-BF1 constructs were normalized by the CAT activity of the cells transfected with the control vector pECE and the respective reporter G5BLCAT2 or BLCAT2. (B) The sequence of a portion of the repression domain in PRDI-BF1. The sequence of the homologous region in Blimp-1 is also shown. The amino acid residues shared by the two proteins are listed between the two sequences. Two stretches of proline-rich regions, PRI and PRII, are indicated by brackets. (C) The proline-rich region of PRDI-BF1 is compared to similar region in WT1. Residues shared by PRDI-BF1 and WT1 protein are listed between the two sequences.
Figure 2
The PRDI-BF1 repression domain functions when fused to the Gal4-DNA-binding domain. (A) The ability of various Gal4–PRDI-BF1 fusion proteins to repress transcription was tested with a reporter that bears five Gal4 DNA-binding sites upstream of the tk promoter (G5BLCAT2). The BLCAT2 (see Fig. 1B) reporter was used as a control. Horizontal lines represent various PRDI-BF1 amino- and carboxy-terminal truncations cloned into the pBXG expression vector, which contains a DNA sequence encoding the Gal4 DNA-binding domain, Gal4(1–147) (boxed). PRDI-BF1 constructs (1 μg) were transfected into HeLa cells with either the control reporter BLCAT2 (dark bars) or G5BLCAT2 (open bars). The data shown are representative of three independent assays, and the CAT activities for all Gal4–PRDI-BF1 constructs were normalized by the CAT activity of the cells transfected with the control vector pECE and the respective reporter G5BLCAT2 or BLCAT2. (B) The sequence of a portion of the repression domain in PRDI-BF1. The sequence of the homologous region in Blimp-1 is also shown. The amino acid residues shared by the two proteins are listed between the two sequences. Two stretches of proline-rich regions, PRI and PRII, are indicated by brackets. (C) The proline-rich region of PRDI-BF1 is compared to similar region in WT1. Residues shared by PRDI-BF1 and WT1 protein are listed between the two sequences.
Figure 3
The PRDI-BF1 protein interacts specifically with the human Groucho-related protein (hGrg) in vitro. (A) Radiolabeled PRDI-BF1 full-length protein or various truncations of PRDI-BF1 were incubated with GST–hGrg protein immobilized on agarose beads. After washing the beads, the bound PRDI-BF1 proteins and one-fifth of the input were analyzed on a 10% SDS–polyacyrlamide gel and visualized by autoradiography. The protein size marker (in kD) is shown at left. (B) Correlation between the repressive function of various PRDI-BF1 truncations and their interaction with GST–hGrg protein is shown. (−) Lack of repression or protein–protein interactions; (+) presence of repressive activity or protein–protein interactions; (n.a.) repressive function was not studied.
Figure 3
The PRDI-BF1 protein interacts specifically with the human Groucho-related protein (hGrg) in vitro. (A) Radiolabeled PRDI-BF1 full-length protein or various truncations of PRDI-BF1 were incubated with GST–hGrg protein immobilized on agarose beads. After washing the beads, the bound PRDI-BF1 proteins and one-fifth of the input were analyzed on a 10% SDS–polyacyrlamide gel and visualized by autoradiography. The protein size marker (in kD) is shown at left. (B) Correlation between the repressive function of various PRDI-BF1 truncations and their interaction with GST–hGrg protein is shown. (−) Lack of repression or protein–protein interactions; (+) presence of repressive activity or protein–protein interactions; (n.a.) repressive function was not studied.
Figure 4
Organization of Groucho family proteins. (A) Domain structures of three forms of the Groucho family proteins. The Q domain is rich in glutamine. The GP domain is proline- and glycine-rich. The CcN domain contains target sites for casein kinase II and cdc2 kinase, and a nuclear localization signal. The SP domain is rich in serine and proline. The carboxy-terminal domain contains four WD-40 repeats. Among these, the Q domain, the CcN domain, and the WD-40 domain are the most conserved. (B) An amino acid sequence alignment of the hGrg, TLE1, TLE2, and Groucho proteins. The entire lengths of hGrg and the amino-terminal 200 amino acids of proteins are shown. Identical residues to hGrg shared by any of the three other proteins are marked by black boxes, and the similar amino acids marked by shaded boxes. Dots denote the alignment gaps.
Figure 5
Groucho family proteins interact with each other and with PRDI-BF1. (A) Diagram of the GST/hGrg, GST/TLE1, and GST/TLE2 fusion constructs used in B–D. The GST domain was fused to various fragments of Groucho proteins, with the numbers indicating the starting and ending amino acids. (B,D) Groucho family proteins TLE1 and TLE2 bind to the repression domain of PRDI-BF1 in vitro. (C) The Q domain of hGrg and TLE1 is sufficient to bind to PRDI-BF1. PRDI-BF1 truncations (lanes 1–22) were translated and radiolabeled in vitro, and incubated with immobilized GST fusion proteins or GST protein, as indicated at the top. After the beads were washed, one-fifth of the input (top) and bound proteins were analyzed on a 10% SDS–polyacrylamide gel and visualized by autoradiography. The protein size marker (in kD) is listed at left. The abnormality of PRDI-BF1(331–789) migration in lane 3 is probably caused by the comigrating GST/TLE2(N) protein, which has a similar molecular weight.
Figure 6
Repression by hGrg and TLE1 proteins fused to the Gal4 DNA-binding domain. (A) Diagram of the Gal4/hGrg and Gal4/TLE1 fusion constructs. (B,C) The full-length hGrg and the amino-terminal Q domains of TLE1 are sufficient for transcriptional repression. HeLa cell transfections similar to those in Fig. 2A were carried out with 0.1 μg of a control vector (pECE) or the indicated Gal4 fusion expression vector, along with a reporter G5BLCAT2 (B) or BLCAT2 (C). Relative CAT activities of the transfections are compared in the histogram.
Figure 6
Repression by hGrg and TLE1 proteins fused to the Gal4 DNA-binding domain. (A) Diagram of the Gal4/hGrg and Gal4/TLE1 fusion constructs. (B,C) The full-length hGrg and the amino-terminal Q domains of TLE1 are sufficient for transcriptional repression. HeLa cell transfections similar to those in Fig. 2A were carried out with 0.1 μg of a control vector (pECE) or the indicated Gal4 fusion expression vector, along with a reporter G5BLCAT2 (B) or BLCAT2 (C). Relative CAT activities of the transfections are compared in the histogram.
Figure 6
Repression by hGrg and TLE1 proteins fused to the Gal4 DNA-binding domain. (A) Diagram of the Gal4/hGrg and Gal4/TLE1 fusion constructs. (B,C) The full-length hGrg and the amino-terminal Q domains of TLE1 are sufficient for transcriptional repression. HeLa cell transfections similar to those in Fig. 2A were carried out with 0.1 μg of a control vector (pECE) or the indicated Gal4 fusion expression vector, along with a reporter G5BLCAT2 (B) or BLCAT2 (C). Relative CAT activities of the transfections are compared in the histogram.
Figure 7
The Q domains of hGrg and TLE1 are sufficient for dimerization. (A) A secondary structure prediction of the entire hGrg sequence using the PHD server. Below the amino acid sequence is the PHD secondary prediction for each residue. (E) Residues likely involved in extended structure (strand); (H) residues involved in forming helix structure; (blank spaces) residues likely to form loop structure or unpredictable. (B) Diagram of the GST–hGrg(N) and GST–TLE1(N2) fusion constructs used in C and D. (C,D) The Q domains of hGrg and TLE1 are sufficient to mediate homo- and heterodimerization. hGrg protein (lanes 1–4) or TLE1 truncations (lanes 5–8) were translated and radiolabeled in vitro, and incubated with immobilized GST–hGrg(N), GST–TLE1(N2) or GST protein, as indicated at the top. After the beads were washed, one-fifth of the input (top) and bound proteins were analyzed on a 10% SDS–polyacrylamide gel and visualized by autoradiography. The protein size marker (in kD) is listed at left.
Figure 8
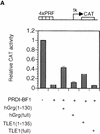
The Q domains of hGrg and TLE1 act as dominant-negative inhibitors of PRDI-BF1 repression. (A,B) Histograms showing CAT activities produced in HeLa cells transfected with a the reporter gene and various hGrg and TLE constructs. One microgram of a control vector (pcDNA3) or a PRDI-BF1 full-length expression vector was cotransfected as indicated by − or +, respectively. One microgram of the expression plasmid containing the Q domain of hGrg [hGrg (1–130)], the full-length hGrg, the Q domain of TLE1 [TLE1(1–135)], or the full-length TLE1 was cotransfected as indicated at the bottom of the graph. (A) Reporter gene containing four tandem PRF sites upstream of the tk promoter driving the CAT gene. (B) A control reporter containing only the tk promoter driving the CAT gene. (C,D) A dominant-negative inhibitor hGrg(1–130) relieves the postinduction repression of the IFN-β promoter. HeLa cells were transfected with 5 μg of a reporter plasmid, indicated at the top, cotransfected with 2 μg of a mammalian expression vector containing [+hGrg(1–130); ▵] or lacking [−hGrg(1–130); ▴] the hGrg(1–130) gene. Twenty-four hours after transfection, the cells were either infected with Sendai virus or untreated (mock induction, █), and SEAP activities in the culture medium were measured at various times following virus infection. (C) Reporter containing the −110 IFN-β promoter driving the expression of the SEAP gene. (D) Reporter containing the herpes virus SV40 promoter driving the SEAP gene (Clontech) (○) −hGrg. The cells were untreated, and SEAP activities were measured at the corresponding time points to those in C.
Figure 8
The Q domains of hGrg and TLE1 act as dominant-negative inhibitors of PRDI-BF1 repression. (A,B) Histograms showing CAT activities produced in HeLa cells transfected with a the reporter gene and various hGrg and TLE constructs. One microgram of a control vector (pcDNA3) or a PRDI-BF1 full-length expression vector was cotransfected as indicated by − or +, respectively. One microgram of the expression plasmid containing the Q domain of hGrg [hGrg (1–130)], the full-length hGrg, the Q domain of TLE1 [TLE1(1–135)], or the full-length TLE1 was cotransfected as indicated at the bottom of the graph. (A) Reporter gene containing four tandem PRF sites upstream of the tk promoter driving the CAT gene. (B) A control reporter containing only the tk promoter driving the CAT gene. (C,D) A dominant-negative inhibitor hGrg(1–130) relieves the postinduction repression of the IFN-β promoter. HeLa cells were transfected with 5 μg of a reporter plasmid, indicated at the top, cotransfected with 2 μg of a mammalian expression vector containing [+hGrg(1–130); ▵] or lacking [−hGrg(1–130); ▴] the hGrg(1–130) gene. Twenty-four hours after transfection, the cells were either infected with Sendai virus or untreated (mock induction, █), and SEAP activities in the culture medium were measured at various times following virus infection. (C) Reporter containing the −110 IFN-β promoter driving the expression of the SEAP gene. (D) Reporter containing the herpes virus SV40 promoter driving the SEAP gene (Clontech) (○) −hGrg. The cells were untreated, and SEAP activities were measured at the corresponding time points to those in C.
Figure 8
The Q domains of hGrg and TLE1 act as dominant-negative inhibitors of PRDI-BF1 repression. (A,B) Histograms showing CAT activities produced in HeLa cells transfected with a the reporter gene and various hGrg and TLE constructs. One microgram of a control vector (pcDNA3) or a PRDI-BF1 full-length expression vector was cotransfected as indicated by − or +, respectively. One microgram of the expression plasmid containing the Q domain of hGrg [hGrg (1–130)], the full-length hGrg, the Q domain of TLE1 [TLE1(1–135)], or the full-length TLE1 was cotransfected as indicated at the bottom of the graph. (A) Reporter gene containing four tandem PRF sites upstream of the tk promoter driving the CAT gene. (B) A control reporter containing only the tk promoter driving the CAT gene. (C,D) A dominant-negative inhibitor hGrg(1–130) relieves the postinduction repression of the IFN-β promoter. HeLa cells were transfected with 5 μg of a reporter plasmid, indicated at the top, cotransfected with 2 μg of a mammalian expression vector containing [+hGrg(1–130); ▵] or lacking [−hGrg(1–130); ▴] the hGrg(1–130) gene. Twenty-four hours after transfection, the cells were either infected with Sendai virus or untreated (mock induction, █), and SEAP activities in the culture medium were measured at various times following virus infection. (C) Reporter containing the −110 IFN-β promoter driving the expression of the SEAP gene. (D) Reporter containing the herpes virus SV40 promoter driving the SEAP gene (Clontech) (○) −hGrg. The cells were untreated, and SEAP activities were measured at the corresponding time points to those in C.
Figure 8
The Q domains of hGrg and TLE1 act as dominant-negative inhibitors of PRDI-BF1 repression. (A,B) Histograms showing CAT activities produced in HeLa cells transfected with a the reporter gene and various hGrg and TLE constructs. One microgram of a control vector (pcDNA3) or a PRDI-BF1 full-length expression vector was cotransfected as indicated by − or +, respectively. One microgram of the expression plasmid containing the Q domain of hGrg [hGrg (1–130)], the full-length hGrg, the Q domain of TLE1 [TLE1(1–135)], or the full-length TLE1 was cotransfected as indicated at the bottom of the graph. (A) Reporter gene containing four tandem PRF sites upstream of the tk promoter driving the CAT gene. (B) A control reporter containing only the tk promoter driving the CAT gene. (C,D) A dominant-negative inhibitor hGrg(1–130) relieves the postinduction repression of the IFN-β promoter. HeLa cells were transfected with 5 μg of a reporter plasmid, indicated at the top, cotransfected with 2 μg of a mammalian expression vector containing [+hGrg(1–130); ▵] or lacking [−hGrg(1–130); ▴] the hGrg(1–130) gene. Twenty-four hours after transfection, the cells were either infected with Sendai virus or untreated (mock induction, █), and SEAP activities in the culture medium were measured at various times following virus infection. (C) Reporter containing the −110 IFN-β promoter driving the expression of the SEAP gene. (D) Reporter containing the herpes virus SV40 promoter driving the SEAP gene (Clontech) (○) −hGrg. The cells were untreated, and SEAP activities were measured at the corresponding time points to those in C.
Similar articles
- Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase.
Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Yu J, et al. Mol Cell Biol. 2000 Apr;20(7):2592-603. doi: 10.1128/MCB.20.7.2592-2603.2000. Mol Cell Biol. 2000. PMID: 10713181 Free PMC article. - Identification and characterization of a novel repressor of beta-interferon gene expression.
Keller AD, Maniatis T. Keller AD, et al. Genes Dev. 1991 May;5(5):868-79. doi: 10.1101/gad.5.5.868. Genes Dev. 1991. PMID: 1851123 - Positive regulatory domain I-binding factor 1 mediates repression of the MHC class II transactivator (CIITA) type IV promoter.
Chen H, Gilbert CA, Hudson JA, Bolick SC, Wright KL, Piskurich JF. Chen H, et al. Mol Immunol. 2007 Feb;44(6):1461-70. doi: 10.1016/j.molimm.2006.04.026. Epub 2006 Jun 12. Mol Immunol. 2007. PMID: 16765445 Free PMC article. - Groucho/TLE family proteins and transcriptional repression.
Chen G, Courey AJ. Chen G, et al. Gene. 2000 May 16;249(1-2):1-16. doi: 10.1016/s0378-1119(00)00161-x. Gene. 2000. PMID: 10831834 Review.
Cited by
- Same rule, different genes: Blimp1 is a pair-rule gene in the milkweed bug Oncopeltus fasciatus.
Reding K, Chung M, Heath A, Hotopp JD, Pick L. Reding K, et al. Sci Adv. 2024 Nov 15;10(46):eadq9045. doi: 10.1126/sciadv.adq9045. Epub 2024 Nov 15. Sci Adv. 2024. PMID: 39546609 Free PMC article. - Spatial microniches of IL-2 combine with IL-10 to drive lung migratory TH2 cells in response to inhaled allergen.
He K, Xiao H, MacDonald WA, Mehta I, Kishore A, Vincent A, Xu Z, Ray A, Chen W, Weaver CT, Lambrecht BN, Das J, Poholek AC. He K, et al. Nat Immunol. 2024 Nov;25(11):2124-2139. doi: 10.1038/s41590-024-01986-8. Epub 2024 Oct 11. Nat Immunol. 2024. PMID: 39394532 - PHLDA1-PRDM1 mediates the effect of lentiviral vectors on fate-determination of human retinal progenitor cells.
Hu X, Chen J, Dai W, Xiao Y, Chen X, Chen Z, Zhang S, Hu Y. Hu X, et al. Cell Mol Life Sci. 2024 Jul 16;81(1):305. doi: 10.1007/s00018-024-05279-z. Cell Mol Life Sci. 2024. PMID: 39012348 Free PMC article. - The transcriptional cofactor Tle3 reciprocally controls effector and central memory CD8+ T cell fates.
Zhao X, Hu W, Park SR, Zhu S, Hu SS, Zang C, Peng W, Shan Q, Xue HH. Zhao X, et al. Nat Immunol. 2024 Feb;25(2):294-306. doi: 10.1038/s41590-023-01720-w. Epub 2024 Jan 18. Nat Immunol. 2024. PMID: 38238608 Free PMC article. - Pioneer and PRDM transcription factors coordinate bivalent epigenetic states to safeguard cell fate.
Matsui S, Granitto M, Buckley M, Ludwig K, Koigi S, Shiley J, Zacharias WJ, Mayhew CN, Lim HW, Iwafuchi M. Matsui S, et al. Mol Cell. 2024 Feb 1;84(3):476-489.e10. doi: 10.1016/j.molcel.2023.12.007. Epub 2024 Jan 10. Mol Cell. 2024. PMID: 38211589
References
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. - PubMed
- Cooper JP, Roth SY, Simpson RT. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes & Dev. 1994;8:1400–1410. - PubMed
- Courey AJ, Huang JD. The establishment and interpretation of transcription factor gradients in the Drosophila embryo. Biochim Biophys Acta. 1995;1261:1–18. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous