Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation - PubMed (original) (raw)
Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation
S J Han et al. Mol Cell Biol. 1999 Feb.
Abstract
The multisubunit Mediator complex of Saccharomyces cerevisiae is required for most RNA polymerase II (Pol II) transcription. The Mediator complex is composed of two subcomplexes, the Rgr1 and Srb4 subcomplexes, which appear to function in the reception of activator signals and the subsequent modulation of Pol II activity, respectively. In order to determine the precise composition of the Mediator complex and to explore the specific role of each Mediator protein, our goal was to identify all of the Mediator components. To this end, we cloned three previously unidentified Mediator subunits, Med9/Cse2, Med10/Nut2, and Med11, and isolated mutant forms of each of them to analyze their transcriptional defects. Differential display and Northern analyses of mRNAs from wild-type and Mediator mutant cells demonstrated an activator-specific requirement for each Mediator subunit. Med9/Cse2 and Med10/Nut2 were required, respectively, for Bas1/Bas2- and Gcn4-mediated transcription of amino acid biosynthetic genes. Gal11 was required for Gal4- and Rap1-mediated transcriptional activation. Med11 was also required specifically for MFalpha1 transcription. On the other hand, Med6 was required for all of these transcriptional activation processes. These results suggest that distinct Mediator proteins in the Rgr1 subcomplex are required for activator-specific transcriptional activation and that the activation signals mediated by these Mediator proteins converge on Med6 (or the Srb4 subcomplex) to modulate Pol II activity.
Figures
FIG. 1
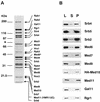
Polypeptide composition of the Mediator-Pol II complex. (A) SDS-polyacrylamide gel electrophoresis of the holopolymerase complex immunopurified on an anti-Rgr1 antibody column. Molecular size marker proteins are indicated on the left, and the Pol II and Mediator components, including the new Mediator proteins (Med9, Med10, and Med11), are indicated on the right. For better resolution of its components, holopolymerase was separated on SDS–7.5% and –13% polyacrylamide gels, and the boundary of the two protein gels is marked with an asterisk. Med6*, histidine-tagged Med6. (B) Immunoblot analysis of the coimmunoprecipitation of the Med proteins. The HA-Med10 holopolymerase fraction was immunoprecipitated with an anti-HA monoclonal antibody (12CA5). Equivalent amounts of load (L), supernatant (S), and pellet (P) from the immunoprecipitation were blotted and probed with the antibodies indicated at the right.
FIG. 2
Sequence alignment of Med10 homologs. Protein sequences of Med10 homologs from S. cerevisiae (GenBank accession no. U25840), S. pombe (2226420), C. elegans (1176601), S. japonicum (AA661070), Arabidopsis thaliana (AA042215), and human (AA429956) are aligned. Identical amino acids (black boxes), similar amino acids (shaded boxes), and gaps in the sequence (dashed line) are indicated.
FIG. 3
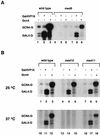
ts phenotypes of the newly identified Mediator mutants. Yeast strains were spotted in duplicate on YPD agar plates, and each plate was incubated for 3 days at either the permissive (30°C) or nonpermissive (37°C) temperature. The temperature sensitivities of the wild-type strains (W), Mediator mutant strains (M), and mutant strains transformed with the corresponding wild-type Mediator gene (rescued [R]) were compared for med9 null (A), med10 ts (B), and med11 ts (C) mutants. At the bottom of each panel, the type of mutations and the mutated amino acids and their positions are shown.
FIG. 4
Northern analysis of genes isolated from differential display of Mediator mutant mRNAs. Wild-type (W) and mutant (M) mRNAs for each of the Mediator genes indicated at the top were prepared from yeast cells grown in YPD at the nonpermissive temperature. Each mRNA blot was hybridized with the probe indicated at the left. As an RNA loading control, actin transcript levels are shown.
FIG. 5
Activator-specific transcriptional defects of Mediator mutants. Poly(A)+ RNA was isolated from wild-type strains (W), mutant strains (M), and mutant strains transformed with the corresponding wild-type Mediator gene (rescued [R]) for each of the Mediator mutants (MED9, MED10, MED11, GAL11, and MED6) noted at the top. In order to measure the levels of the transcripts indicated at the left, cells were grown under galactose induction (GAL1), amino acid-rich conditions (HIS4 B), or amino acid starvation conditions (HIS4 A). All of the yeast cells used in this experiment are α mating type cells and thus display activated MFα1 transcription (MFα1). The levels of MFα1 transcripts in the med9 and med11 mutant strains containing then wild-type Mediator genes (MED9 R and MED11 R) and in the gal11 mutant strain (GAL11 W and M) were not determined. Each blot was hybridized with the probes indicated at the left. For each Northern blot, the level of actin mRNA was measured as an RNA loading control, and a typical result is shown.
FIG. 6
Activator-specific requirement for Med9 and Med10. The structures of the lacZ reporter constructs containing a binding site(s) for one activator type are shown along with the transcriptional activator used for the assay. The β-galactosidase activities from the averages of two triplicate assays are shown with standard deviations (SD). The strains used were YPHBAS (wild type1), YSJ9BAS med9 null (_med9_1), and YSJ10BAS med10 ts (_med10_1) for the Bas2 assay, while YPHG4 (wild type2), YSJ9G4 med9 null (_med9_2), and YSJ10G4 med10 ts (_med10_2) were used for the Gcn4 assay.
FIG. 7
In vitro transcription activities of Mediator mutant holopolymerases. In vitro transcription reactions were reconstituted as described by Lee et al. (24). Activators (Gal4VP16 [30 ng] and Gcn4 [30 ng]) were added to the reaction mixtures as indicated. The reaction mixtures were incubated on ice (A) or at the indicated temperature (B) for 10 min for initiation complex formation, and then [α-32P]UTP (10 μCi) and 0.5 mM CTP were added and the reaction mixtures were incubated further at 25°C for 30 min. Specifically initiated transcripts from a template containing either a Gcn4 binding site (GCN4:G-) or a Gal4 binding site (GAL:G-) are indicated. (A) Transcriptional activity of wild-type holopolymerase (lanes 1 to 3) and equivalent amounts of med9 null holopolymerase based on nonspecific RNA polymerase activity (lanes 4 to 6) or based on Mediator content (lanes 7 to 9). (B) Transcriptional activity of wild-type (lanes 1 to 3 and 10 to 12), med10 ts (lanes 4 to 6 and 13 to 15), and med11 ts (lanes 7 to 9 and 16 to 18) holopolymerases under permissive (25°C, lanes 1 to 9) or restrictive (37°C, lanes 10 to 18) conditions.
FIG. 8
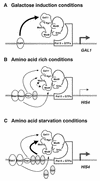
A model for activator-specific modules of Mediator complex. Three models for activator-specific interactions of the Mediator complex are shown. Filled and linear arrows indicate the specific functional interactions revealed by in vivo transcription assays. Shaded arrows represent transcription initiation. (A) Galactose induction conditions. Gal4 bound to the enhancer interacts with the Gal11 module of Mediator, which induces Pol II and the general transcription factors (GTFs) to transcribe GAL1 at higher efficiency. (B) Amino acid-rich conditions. Med9 and Med10 mediate specifically basal-level HIS4 transcription via Bas1/Bas2 and Gcn4, respectively. Stimulation of Bas1/Bas2- and Gcn4-dependent transcription by Rap1 through the Gal11 module is indicated by a linear arrow. The putative requirement for Med10 in Bas2-mediated transcription is indicated by a dotted arrow. (C) Amino acid starvation conditions. The major transcriptional activation of HIS4 by the specific interaction of (overproduced) Gcn4 protein with Med10 is shown as a filled arrow, compared to the relatively minor contribution from Rap1 to the Gcn4-mediated transcriptional activation via the Gal11 module (linear arrow).
Similar articles
- Med9/Cse2 and Gal11 modules are required for transcriptional repression of distinct group of genes.
Han SJ, Lee JS, Kang JS, Kim YJ. Han SJ, et al. J Biol Chem. 2001 Oct 5;276(40):37020-6. doi: 10.1074/jbc.M105596200. Epub 2001 Jul 24. J Biol Chem. 2001. PMID: 11470794 - Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription.
Lee YC, Kim YJ. Lee YC, et al. Mol Cell Biol. 1998 Sep;18(9):5364-70. doi: 10.1128/MCB.18.9.5364. Mol Cell Biol. 1998. PMID: 9710620 Free PMC article. - An activator binding module of yeast RNA polymerase II holoenzyme.
Lee YC, Park JM, Min S, Han SJ, Kim YJ. Lee YC, et al. Mol Cell Biol. 1999 Apr;19(4):2967-76. doi: 10.1128/MCB.19.4.2967. Mol Cell Biol. 1999. PMID: 10082564 Free PMC article. - Mediator of transcriptional regulation.
Myers LC, Kornberg RD. Myers LC, et al. Annu Rev Biochem. 2000;69:729-49. doi: 10.1146/annurev.biochem.69.1.729. Annu Rev Biochem. 2000. PMID: 10966474 Review. - The yeast mediator.
Björklund S, Buzaite O, Hallberg M. Björklund S, et al. Mol Cells. 2001 Apr 30;11(2):129-36. Mol Cells. 2001. PMID: 11355691 Review.
Cited by
- A novel tRNA-derived fragment AS-tDR-007333 promotes the malignancy of NSCLC via the HSPB1/MED29 and ELK4/MED29 axes.
Yang W, Gao K, Qian Y, Huang Y, Xiang Q, Chen C, Chen Q, Wang Y, Fang F, He Q, Chen S, Xiong J, Chen Y, Xie N, Zheng D, Zhai R. Yang W, et al. J Hematol Oncol. 2022 May 7;15(1):53. doi: 10.1186/s13045-022-01270-y. J Hematol Oncol. 2022. PMID: 35526007 Free PMC article. - The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II.
Bhoite LT, Yu Y, Stillman DJ. Bhoite LT, et al. Genes Dev. 2001 Sep 15;15(18):2457-69. doi: 10.1101/gad.921601. Genes Dev. 2001. PMID: 11562354 Free PMC article. - MED29, a component of the mediator complex, possesses both oncogenic and tumor suppressive characteristics in pancreatic cancer.
Kuuselo R, Savinainen K, Sandström S, Autio R, Kallioniemi A. Kuuselo R, et al. Int J Cancer. 2011 Dec 1;129(11):2553-65. doi: 10.1002/ijc.25924. Epub 2011 Mar 25. Int J Cancer. 2011. PMID: 21225629 Free PMC article. - Core Mediator structure at 3.4 Å extends model of transcription initiation complex.
Nozawa K, Schneider TR, Cramer P. Nozawa K, et al. Nature. 2017 May 11;545(7653):248-251. doi: 10.1038/nature22328. Epub 2017 May 3. Nature. 2017. PMID: 28467824 - Toward understanding of the mechanisms of Mediator function in vivo: Focus on the preinitiation complex assembly.
Eychenne T, Werner M, Soutourina J. Eychenne T, et al. Transcription. 2017;8(5):328-342. doi: 10.1080/21541264.2017.1329000. Epub 2017 Aug 25. Transcription. 2017. PMID: 28841352 Free PMC article. Review.
References
- Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. - PubMed
- Bjorklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. - PubMed
- Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. - PubMed
- Chen X H, Xiao Z, Fitzgerald-Hayes M. SCM2, a tryptophan permease in Saccharomyces cerevisiae, is important for cell growth. Mol Gen Genet. 1994;244:260–268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases