A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development - PubMed (original) (raw)
A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development
J Nielsen et al. Mol Cell Biol. 1999 Feb.
Abstract
Insulin-like growth factor II (IGF-II) is a major fetal growth factor. The IGF-II gene generates multiple mRNAs with different 5' untranslated regions (5' UTRs) that are translated in a differential manner during development. We have identified a human family of three IGF-II mRNA-binding proteins (IMPs) that exhibit multiple attachments to the 5' UTR from the translationally regulated IGF-II leader 3 mRNA but are unable to bind to the 5' UTR from the constitutively translated IGF-II leader 4 mRNA. IMPs contain the unique combination of two RNA recognition motifs and four hnRNP K homology domains and are homologous to the Xenopus Vera and chicken zipcode-binding proteins. IMP localizes to subcytoplasmic domains in a growth-dependent and cell-specific manner and causes a dose-dependent translational repression of IGF-II leader 3 -luciferase mRNA. Mouse IMPs are produced in a burst at embryonic day 12.5 followed by a decline towards birth, and, similar to IGF-II, IMPs are especially expressed in developing epithelia, muscle, and placenta in both mouse and human embryos. The results imply that cytoplasmic 5' UTR-binding proteins control IGF-II biosynthesis during late mammalian development.
Figures
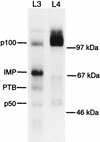
FIG. 1
UV cross-linking of IGF-II leaders 3 and 4 to an RD cytoplasmic extract. An autoradiograph from a UV cross-linking analysis of human IGF-II leader 3 (L3) and leader 4 (L4) in the presence of a detergent-solubilized RD cytoplasmic extract is shown. Cross-linked species and molecular size markers are indicated on the left and right, respectively.
FIG. 2
Modular structure of the human IMP family. (A) Alignment of three members of the IMP family, the zipcode-binding protein (ZBP), and the Vera protein, with black and grey boxes indicating amino acid identity and similarity, respectively. Horizontal lines below the alignment indicate the predicted two RRMs and four KH domains. (B) Schematic diagram of RRMs and KH domains indicated in panel A.
FIG. 3
Segments of IGF-II leader 3 RNA bind IMP. (A) Map of the RNA segments obtained from leader 3. Numbers below the map refer to the numbering of human IGF-II exon 5 containing 1,164 nucleotides of leader 3 RNA (31). (B) Mobility shift analysis of leader 3 RNA segments and full-length leader 4 RNA with recombinant IMP-1. In experiments with the four segments A to D, the concentrations of recombinant IMP-1 are 0, 0.07, 0.3, and 1 nM, respectively, whereas 0 and 10 nM are used in the leader 4 experiment. (C) UV cross-linking of full-length leader 3 RNA (L3) and segments A to D to a detergent-solubilized RD cytoplasmic extract. Cross-linked species and molecular size markers are indicated on the left and right, respectively.
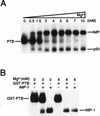
FIG. 4
Mg2+-dependent competition of PTB and IMP. (A) UV cross-linking of full-length leader 3 RNA to a detergent-solubilized RD cytoplasmic extract in the presence of increasing Mg2+ concentrations. The major cross-linked species are indicated to the left if abundant at a low Mg2+ concentration and to the right if preferred at a high Mg2+ concentration. Moreover, the putative p50 major core protein (6) is indicated. (B) UV cross-linking of segment C to recombinant GST-PTB (8 nM) and IMP-1 (8 nM) at 0 and 4 mM Mg2+. Lanes 2 and 5 are the result of UV cross-linking in the presence of both proteins at 0 and 4 mM Mg2+, respectively.
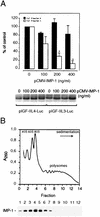
FIG. 5
IMP-1 inhibits translation of a chimeric IGF-II leader 3-luciferase transcript. (A) NIH 3T3 cells were transiently transfected with pIGF-IIL4-Luc or pIGF-IIL3-Luc reporter constructs and increasing concentrations of pCMV-IMP-1 as indicated. The graph depicts the dose-response relationship between the amount of pCMV-IMP-1 and the obtained luciferase activity. The results are given as a percentage of control (mean and standard error of the mean of two to four independent experiments), and the asterisks indicate statistically significant differences at a P of <0.05. A representative Northern analysis indicating the levels of pIGF-IIL4-Luc or pIGF-IIL3-Luc reporter mRNAs is shown below the bar graph. (B) Subcellular distribution of IMP-1. A detergent-solubilized RD cytoplasmic extract was applied to a linear 20 to 47% sucrose gradient. The graph shows the _A_260 profile of the sedimenting lysate. The positions of the 40S, 60S, 80S, and polysomal complexes are indicated. Fractions were precipitated in 10% trichloroacetic acid, and proteins were separated by SDS-polyacrylamide gel electrophoresis, followed by Western analysis with an anti-IMP-1 antibody (lower panel).
FIG. 6
Subcellular localization of IMP-1. The subcellular distribution of IMP-1 was characterized immunocytochemically. RD or NIH 3T3 cells were transiently transfected with pCMV-IMP-1. After 48 h, the cells were fixed and incubated with rabbit anti-IMP-1 and stained with fluorescein isothiocyanate-conjugated anti-rabbit IgG. (a to c) Subcellular distribution of IMP-1 in RD cells; (d to f) distribution of IMP-1 in NIH 3T3 cells. (a) In RD cells, staining was observed below the plasma membrane. (b) In clustered groups of cells, IMP-1 was polarized to the free edges of the cells; (c) in myotubes, staining was distributed along the circumference of the complete myotube. (d) In NIH 3T3 cells, staining was evenly distributed in the cytoplasm in clustered cells. (e) In areas with low cell density, staining was observed in the lamellipodia of the leading edge and in the perinuclear region. (f) In completely dispersed cells, IMP-1 was found in discrete foci scattered around the nucleus, below the plasma membrane, and in the lamellipodia. Magnification, ×1,000. Arrows in panels a and d indicate nontransfected cells that served as negative controls for the immunostaining. See Materials and Methods for additional controls.
FIG. 7
Expression of IMP-1, IMP-2, and IMP-3 in mouse and human embryos. (A) Expression of mouse equivalents of IMP-1, IMP-2, and IMP-3 mRNAs was examined by Northern analysis of total RNA from different embryonic (8.5 to 17.5) and postnatal (100) days as indicated. The amount of RNA in each lane is normalized to rRNA content. (B) Western analysis of IMP-1 in mouse embryos at E12.5, E14.5, and E17.5. The amounts of loaded protein in the three lanes are identical, and lane C contains a positive control sample. (C) Immunohistochemical staining of IMP-1 in mouse and human embryos. (a and b) Localization of IMP-1 in mouse epidermis at E12.5. (a) Staining is visible in the basal layer of the developing epidermis at the basal plasma cell membrane, as indicated by the arrow in panel b. (c) Parallel section where the antiserum was preadsorbed with the peptide used for immunization. (d) Localization of IMP-1 in developing mouse muscle at E15.5. (e) Localization of IMP-1 in fetal human skeletal muscle cells (38 weeks). The submembranous staining is indicated by an arrow. (f) Staining of human term placenta shows the expression of IMP-1 in the trophoblast cells. Scale bars, 11 (a), 7 (b), 11 (c), 20 (d), 11 (e), and 11 (f) μm.
References
- Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. - PubMed
- Baker J, Liu J P, Robertson E J, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. - PubMed
- Brice A L, Cheetham J E, Bolton V N, Hill N C, Schofield P N. Temporal changes in the expression of the insulin-like growth factor II gene associated with tissue maturation in the human fetus. Development. 1989;106:543–554. - PubMed
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases